BAQALC: Blockchain Applied Lossless Efficient Transmission of DNA Sequencing Data for Next Generation Medical Informatics
Abstract
:1. Introduction
2. Background
2.1. DNA Data Composition
2.2. Data Compression
2.3. Prior Research on DNA Compression
2.4. Blockchain Technology
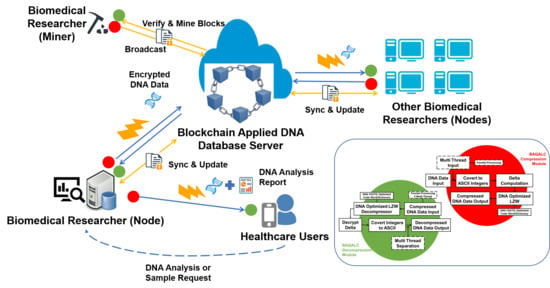
3. System Description
3.1. Overall Architecture
3.2. Proposed Solution: BAQALC Algorithm
4. Results
4.1. Materials and Methods
4.2. Comparison between Datasets
4.3. Compression Ratio Comparison to Widely Used Algorithms
4.4. Compression Ratio Comparison to Related Research
4.5. Overall CR and Stability Comparison
5. Discussion and Conclusions
Supplementary Files
Supplementary File 1Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Mei, X.P.; Gao, C.J.; Zhang, G.H.; Sun, X.D. Histologic Distribution, Fragment Cloning, and Sequence Analysis of G Protein Couple Receptor 30 in Rat Submaxillary Gland. Anat. Rec. Integr. Anat. Evol. Biol. 2011, 294, 706–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sardaraz, M.; Tahir, M.; Ikram, A.A. Advances in high throughput DNA sequence data compression. J. Bioinf. Comput. Biol. 2016, 14, 1630002. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, Y.; Ji, Z.; He, S.; Yang, X. High-throughput DNA sequence data compression. Briefings Bioinf. 2015, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Showell, C. Barriers to the use of personal health records by patients: A structured review. PeerJ 2017, 5, e3268. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Cho, G.Y.; Song, S.H.; Jang, J.S.; Lee, K.I.; Lee, T.R. Solution for Efficient Vital Data Transmission and Storing in m-Health Environment. J. Digit. Converg. 2015, 13, 227–235. [Google Scholar] [CrossRef]
- Bouillaguet, C.; Derbez, P.; Dunkelman, O.; Keller, N.; Rijmen, V.; Fouque, P.A. Low-data complexity attacks on AES. IEEE Trans. Inf. Theory 2012, 58, 7002–7017. [Google Scholar] [CrossRef]
- Zhang, L.Y.; Liu, Y.; Wang, C.; Zhou, J.; Zhang, Y.; Chen, G. Improved known-plaintext attack to permutation-only multimedia ciphers. Inf. Sci. 2018, 430–431, 228–239. [Google Scholar] [CrossRef]
- Hosseini, M.; Pratas, D.; Pinho, A.J. Cryfa: A secure encryption tool for genomic data. Bioinformatics 2018, bty645. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.C.; Ruzzo, W.L.; Peng, X.; Katze, M.G. Compression of next-generation sequencing reads aided by highly efficient de novo assembly. Nucleic Acids Res. 2012, 40, e171. [Google Scholar] [CrossRef] [PubMed]
- Tembe, W.; Lowey, J.; Suh, E. G-SQZ: Compact encoding of genomic sequence and quality data. Bioinformatics 2010, 26, 2192–2194. [Google Scholar] [CrossRef] [PubMed]
- Hach, F.; Numanagic, I.; Sahinalp, S.C. DeeZ: Reference-based compression by local assembly. Nat. Methods 2014, 11, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Salah, K. IoT security: Review, blockchain solutions, and open challenges. Future Gener. Comput. Syst. 2018, 82, 395–411. [Google Scholar] [CrossRef]
- Lee, S.J.; Rho, M.J.; Yook, I.H.; Park, S.H.; Jang, K.S.; Park, B.J.; Lee, O.; Lee, D.J.; Choi, I.Y. Design, Development and Implementation of a Smartphone Overdependence Management System for the Self-Control of Smart Devices. Appl. Sci. 2016, 6. [Google Scholar] [CrossRef]
- Doolittle, G.C.; Spaulding, A.O.; Williams, A.R. The Decreasing Cost of Telemedicine and Telehealth. Telemed. J. E Health 2011, 17, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Gonzalez, S.; Leung, V.; Zhang, Q.; Li, M. A 2G-RFID-Based E-Healthcare System. IEEE Wirel. Commun. 2010, 17, 37–43. [Google Scholar] [CrossRef]
- What is DNA?–Genetics Home Reference–NIH. Available online: https://ghr.nlm.nih.gov/primer/basics/dna (accessed on 23 August 2018).
- Bonfield, J.K.; Mahoney, M.V. Compression of FASTQ and SAM format sequencing data. PLoS ONE 2013, 8, e59190. [Google Scholar] [CrossRef] [PubMed]
- Guerra, A.; Lotero, J.; Isaza, S. Performance comparison of sequential and parallel compression applications for DNA raw data. J. Supercomput. 2016, 72, 4696–4717. [Google Scholar] [CrossRef]
- Cho, G.Y.; Lee, S.J.; Lee, T.R. An optimized compression algorithm for real-time ECG data transmission in wireless network of medical information systems. J. Med. Syst. 2015, 39, 161. [Google Scholar] [CrossRef] [PubMed]
- Cho, G.Y.; Lee, G.Y.; Lee, T.R. Efficient Real-Time Lossless EMG Data Transmission to Monitor Pre-Term Delivery in a Medical Information System. Appl. Sci. 2017, 7. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, G.; Jiang, H.; Meng, S. Research and improvement of ECG compression algorithm based on EZW. Comput. Methods Programs Biomed. 2017, 145, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wu, T.; Wan, Z.; Song, Z.; Yu, M.; Wang, D.; Li, L.; Chen, F.; Xu, X. A method to differentiate between ventricular fibrillation and asystole during chest compressions using artifact-corrupted ECG alone. Comput. Methods Programs Biomed. 2017, 141, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Hach, F.; Numanagic, I.; Alkan, C.; Sahinalp, S.C. SCALCE: Boosting sequence compression algorithms using locally consistent encoding. Bioinformatics 2012, 28, 3051–3057. [Google Scholar] [CrossRef] [PubMed]
- Ziv, J.; Lempel, A. Compression of Individual Sequences via Variable-Rate Coding. IEEE Trans. Inf. Theory 1978, 24, 530–536. [Google Scholar] [CrossRef]
- Ziv, J.; Lempel, A. Universal Algorithm for Sequential Data Compression. IEEE Trans. Inf. Theory 1977, 23, 337–343. [Google Scholar] [CrossRef]
- Roguski, L.; Deorowicz, S. DSRC 2--Industry-oriented compression of FASTQ files. Bioinformatics 2014, 30, 2213–2215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanford, K.I.; Middelbeek, R.J.; Goodyear, L.J. Exercise Effects on White Adipose Tissue: Beiging and Metabolic Adaptations. Diabetes 2015, 64, 2361–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocyte. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.A.; Wen, Z.; Deng, Q.; Chu, Y.; Sun, Y.; Zhu, Z. LW-FQZip 2: A parallelized reference-based compression of FASTQ files. BMC Bioinf. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Fritz, M.H.Y.; Leinonen, R.; Cochrane, G.; Birney, E. Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res. 2011, 21, 734–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolae, M.; Pathak, S.; Rajasekaran, S. LFQC: A lossless compression algorithm for FASTQ files. Bioinformatics 2015, 31, 3276–3281. [Google Scholar] [CrossRef] [PubMed]
- Al-Okaily, A.; Almarri, B.; Al Yami, S.; Huang, C.H. Toward a Better Compression for DNA Sequences Using Huffman Encoding. J. Comput. Biol. 2017, 24, 280–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Gzip Homepage. Available online: https://www.gzip.org/ (accessed on 23 August 2018).
- Pinho, A.J.; Pratas, D. MFCompress: A compression tool for FASTA and multi-FASTA data. Bioinformatics 2014, 30, 117–118. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.H.; Dutta, A.; Bost, T.; Chadaram, S. DELIMINATE—A fast and efficient method for loss-less compression of genomic sequences. Bioinformatics 2012, 28, 2527–2529. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chen, X.; Xiang, Y. Blockchain-based publicly verifiable data deletion scheme for cloud storage. J. Netw. Comput. Appl. 2018, 103, 185–193. [Google Scholar] [CrossRef]
- Goni, A.; Burgos, A.; Dranca, L.; Rodriguez, J.; Illarramendi, A.; Bermudez, J. Architecture, cost-model and customization of real-time monitoring systems based on mobile biological sensor data-streams. Comput. Methods Programs Biomed. 2009, 96, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, R.; Sugawara, H.; Shumway, M. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- The Top 10 Causes of Death, Fact Sheets. Available online: http://www.who.int/mediacentre/factsheets/fs310/en/index1.html (accessed on 23 August 2018).
- Ischemic Cardiomyopathy: Symptoms, Causes, and Treatment. Available online: https://www.healthline.com/health/ischemic-cardiomyopathy (accessed on 23 August 2018).
- Friedrichs, I.; Bingold, T.; Keppler, O.T.; Pullmann, B.; Reinheimer, C.; Berger, A. Detection of herpesvirus EBV DNA in the lower respiratory tract of ICU patients: A marker of infection of the lower respiratory tract? Med. Microbiol. Immunol. 2013, 202, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, S.A.; Scoggin, C.H. DNA molecular biology in the diagnosis of pulmonary disease. Clin. Chest Med. 1987, 8, 161–171. [Google Scholar] [PubMed]
- Taneja, T.K.; Sharma, S.K. Markers of small cell lung cancer. World J Surg. Oncol. 2004, 2, 10. [Google Scholar] [CrossRef] [PubMed]





| Solution | Contents |
|---|---|
| LW-FQZip 2 [31] | Parallelized reference-based compression of FASTQ files. Tool for archival or space-sensitive (quality scores, metadata, and nucleotide bases) applications of high-throughput DNA sequence data. |
| QUIP [11] | Lossless compression algorithm. Adopted a reference-based compression, and known as the first assembly-based compressor that uses a novel de novo assembly algorithm. |
| DSRC 2 [28] | In this algorithm, a single thread reads the input FASTQ file in blocks (typically of tens of MBs size) and puts them into an input queue. Several threads perform the compression of the blocks, storing the results in an output queue. Finally, a single thread writes the compressed blocks in a single file. |
| CRAM [32] | Reference-based compression method. Targets well-studied genomes. Aligns new sequences to a reference genome and then encodes the differences between the new sequence and the reference genome for storage. |
| LFQC [33] | Lossless non-reference based FASTQ compression algorithm by generating identifier fields systematically. |
| SCALCE [25] | Boosting scheme based on locally consistent parsing technique, which reorganizes the DNA reads in a way that results in a higher compression speed and compression rate, independent of the compression algorithm in use and without using a reference genome. |
| UHT [34] | DNA compression based on using Huffman coding. Unbalanced Huffman encoding/Tree, forcing the Huffman tree to be unbalanced to be better than the standard Huffman. |
| UHTL [34] | DNA compression based on using Huffman coding. Developed version of UHT that prioritizes encoding the k-mers that contain the least frequent base. |
| MUHTL [34] | DNA compression based on using Huffman coding. Developed version of UHTL that allows more k-mers to be encoded to apply multiple Standard Huffman Tree (SHT)/UHT/UHTL coding. |
| FQZCOMP [19] | Proposed Fqzcomp and Fastqz that both accept FASTQ files as input, with the latter also taking an optional genome sequence to perform reference-based compression. Additionally proposes Samcomp, which also performs reference-based compression but requires previously aligned data in the SAM format instead. |
| Database | CR | Used Data | CR Average ± SD |
|---|---|---|---|
| Insulin | 10.98 | SRX2528043 | 11.17 ± 0.11 |
| 11.24 | SRX2528042 | ||
| 11.22 | SRX2528041 | ||
| 11.25 | SRX2528040 | ||
| 11.18 | SRX2528039 | ||
| Epstein-Barr | 14.7 | SRX2776811 | 13.65 ± 0.7 |
| 13.79 | SRX2776810 | ||
| 13.63 | SRX2776809 | ||
| 13.36 | SRX2776808 | ||
| 12.78 | SRX2776807 | ||
| Lung | 17.01 | SRX2388195 | 17.01 ± 0.06 |
| 16.98 | SRX2388194 | ||
| 17.08 | SRX2388193 | ||
| 17.06 | SRX2388192 | ||
| 16.94 | SRX2388191 | ||
| Leptin | 10 | ERX1600597 | 10.61 ± 0.56 |
| 10.25 | ERX1600596 | ||
| 10.44 | ERX1600595 | ||
| 11 | ERX1600594 | ||
| 11.38 | ERX1600593 | ||
| Cystic Fibrosis | 10.06 | SRX2935589 | 10.09 ± 0.15 |
| 10.13 | SRX2935588 | ||
| 9.85 | SRX2935587 | ||
| 10.17 | SRX2935586 | ||
| 10.26 | SRX2935585 | ||
| Total CR | All Data | 12.51 ± 2.64 | |
| Solutions | CR | Used Data | CR Average ± SD |
|---|---|---|---|
| SHT | 2.22 | SRX2935589 | 2.67 ± 0.72 |
| 2.22 | SRX2935588 | ||
| 2.22 | SRX2935587 | ||
| 2.22 | SRX2935586 | ||
| 2.22 | SRX2935585 | ||
| 2.2 | SRX2528043 | ||
| 2.21 | SRX2528042 | ||
| 2.21 | SRX2528041 | ||
| 2.21 | SRX2528040 | ||
| 2.21 | SRX2528039 | ||
| 3.44 | Cholerae | ||
| 3.44 | Abscessus | ||
| 3.34 | S. cerevisiae | ||
| 3.27 | N. crassa | ||
| 4.45 | Chr22 | ||
| Gzip | 3.38 | SRR2916693 | 3.41 ± 0.87 |
| 2.92 | SRR2994368 | ||
| 2.35 | SRR3211986 | ||
| 2.2 | ERR739513 | ||
| 3.39 | SRR3190692 | ||
| 5.59 | ERR385912 | ||
| 3.85 | ERR386131 | ||
| 2.71 | SRR034509 | ||
| 3.15 | ERR174310 | ||
| 4.24 | ERR194147 | ||
| 3.29 | Cholerae | ||
| 3.31 | Abscessus | ||
| 3.1 | S. cerevisiae | ||
| 3.12 | N. crassa | ||
| 4.57 | Chr22 | ||
| BAQALC | 11.22 | SRX2528041 | 12.51 ± 2.58 |
| 11.25 | SRX2528040 | ||
| 11.18 | SRX2528039 | ||
| 10.44 | ERX1600595 | ||
| 11 | ERX1600594 | ||
| 11.38 | ERX1600593 | ||
| 13.63 | SRX2776809 | ||
| 13.36 | SRX2776808 | ||
| 12.78 | SRX2776807 | ||
| 9.85 | SRX2935587 | ||
| 10.17 | SRX2935586 | ||
| 10.26 | SRX2935585 | ||
| 17.08 | SRX2388193 | ||
| 17.06 | SRX2388192 | ||
| 16.94 | SRX2388191 | ||
| LZW | 3.24 | SRX2935588 | 3.24 ± 0.12 |
| 3.23 | SRX2935587 | ||
| 3.24 | SRX2935586 | ||
| 3.24 | SRX2935585 | ||
| 3.21 | SRX2528043 | ||
| 3.21 | SRX2528042 | ||
| 2.88 | SRX2528041 | ||
| 3.21 | SRX2528040 | ||
| 3.2 | SRX2528039 | ||
| 3.2 | ERX1600597 | ||
| 3.38 | ERX1600596 | ||
| 3.35 | ERX1600595 | ||
| 3.34 | ERX1600594 | ||
| 3.32 | ERX1600593 | ||
| 3.31 | SRX2388195 | ||
| Bzip 2 | 4.13 | SRR2916693 | 3.99 ± 1.15 |
| 3.51 | SRR2994368 | ||
| 2.75 | SRR3211986 | ||
| 2.52 | ERR739513 | ||
| 4.1 | SRR3190692 | ||
| 7.19 | ERR385912 | ||
| 4.65 | ERR386131 | ||
| 3.17 | SRR034509 | ||
| 3.82 | ERR174310 | ||
| 5.08 | ERR194147 | ||
| 3.52 | Cholerae | ||
| 3.57 | Abscessus | ||
| 3.41 | S. cerevisiae | ||
| 3.43 | N. crassa | ||
| 5.04 | Chr22 |
| Researches | CR | Used Data | CR Average ± SD |
|---|---|---|---|
| LW-FQZip 2 | 6.54 | SRR2916693 | 6.99 ± 4.9 |
| 6.25 | SRR2994368 | ||
| 3.1 | SRR3211986 | ||
| 2.87 | ERR739513 | ||
| 8.55 | SRR3190692 | ||
| 20.0 | ERR385912 | ||
| 6.25 | ERR386131 | ||
| 4.41 | SRR034509 | ||
| 4.98 | ERR174310 | ||
| 6.99 | ERR194147 | ||
| DSRC | 4.95 | SRR2916693 | 5.83 ± 2.89 |
| 4.31 | SRR2994368 | ||
| 4.93 | SRR3190692 | ||
| 12.82 | ERR385912 | ||
| 5.95 | ERR386131 | ||
| 3.83 | SRR034509 | ||
| 4.95 | ERR174310 | ||
| 4.93 | ERR194147 | ||
| LFQC | 7.87 | SRR2916693 | 6.96 ± 5.4 |
| 3.1 | SRR3211986 | ||
| 2.87 | ERR739513 | ||
| 17.24 | ERR385912 | ||
| 6.45 | ERR386131 | ||
| 4.22 | SRR034509 | ||
| UHT | 3.69 | Cholerae | 3.91 ± 0.5 |
| 3.74 | Abscessus | ||
| 3.66 | S. cerevisiae | ||
| 3.68 | N. crassa | ||
| 4.8 | Chr22 | ||
| MUHTL | 3.85 | Cholerae | 4.09 ± 0.55 |
| 3.91 | Abscessus | ||
| 3.83 | S. cerevisiae | ||
| 3.8 | N. crassa | ||
| 5.07 | Chr22 | ||
| Quip | 4.78 | SRR2916693 | 5.82 ± 3.16 |
| 4.98 | SRR2994368 | ||
| 3.0 | SRR3211986 | ||
| 6.06 | SRR3190692 | ||
| 13.89 | ERR385912 | ||
| 5.65 | ERR386131 | ||
| 3.98 | SRR034509 | ||
| 5.0 | ERR174310 | ||
| 5.0 | ERR194147 | ||
| CRAM | 4.57 | SRR2916693 | 3.74 ± 0.68 |
| 3.79 | SRR2994368 | ||
| 2.95 | SRR3211986 | ||
| 2.81 | ERR739513 | ||
| 4.48 | SRR3190692 | ||
| 3.92 | ERR386131 | ||
| 3.65 | SRR034509 | ||
| SCALCE | 5.81 | SRR2916693 | 6.59 ± 3.5 |
| 5.78 | SRR2994368 | ||
| 2.99 | SRR3211986 | ||
| 7.87 | SRR3190692 | ||
| 15.15 | ERR385912 | ||
| 6.02 | ERR386131 | ||
| 4.08 | SRR034509 | ||
| 5.1 | ERR174310 | ||
| 6.49 | ERR194147 | ||
| UHTL | 3.84 | Cholerae | 4.08 ± 0.55 |
| 3.89 | Abscessus | ||
| 3.83 | S. cerevisiae | ||
| 3.8 | N. crassa | ||
| 5.06 | Chr22 | ||
| FQZCOMP | 4.69 | SRR003177 | 5.95 ± 1.31 |
| 7.04 | SRR007215_1 | ||
| 4.6 | SRR027520_1 | ||
| 7.46 | SRR065390_1 | ||
| BAQALC | 10.98 | SRX2528043 | 12.51 ± 2.86 |
| 11.24 | SRX2528042 | ||
| 10 | ERX1600597 | ||
| 10.25 | ERX1600596 | ||
| 14.7 | SRX2776811 | ||
| 13.79 | SRX2776810 | ||
| 10.06 | SRX2935589 | ||
| 10.13 | SRX2935588 | ||
| 17.01 | SRX2388195 | ||
| 16.98 | SRX2388194 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.-J.; Cho, G.-Y.; Ikeno, F.; Lee, T.-R. BAQALC: Blockchain Applied Lossless Efficient Transmission of DNA Sequencing Data for Next Generation Medical Informatics. Appl. Sci. 2018, 8, 1471. https://doi.org/10.3390/app8091471
Lee S-J, Cho G-Y, Ikeno F, Lee T-R. BAQALC: Blockchain Applied Lossless Efficient Transmission of DNA Sequencing Data for Next Generation Medical Informatics. Applied Sciences. 2018; 8(9):1471. https://doi.org/10.3390/app8091471
Chicago/Turabian StyleLee, Seo-Joon, Gyoun-Yon Cho, Fumiaki Ikeno, and Tae-Ro Lee. 2018. "BAQALC: Blockchain Applied Lossless Efficient Transmission of DNA Sequencing Data for Next Generation Medical Informatics" Applied Sciences 8, no. 9: 1471. https://doi.org/10.3390/app8091471
APA StyleLee, S. -J., Cho, G. -Y., Ikeno, F., & Lee, T. -R. (2018). BAQALC: Blockchain Applied Lossless Efficient Transmission of DNA Sequencing Data for Next Generation Medical Informatics. Applied Sciences, 8(9), 1471. https://doi.org/10.3390/app8091471