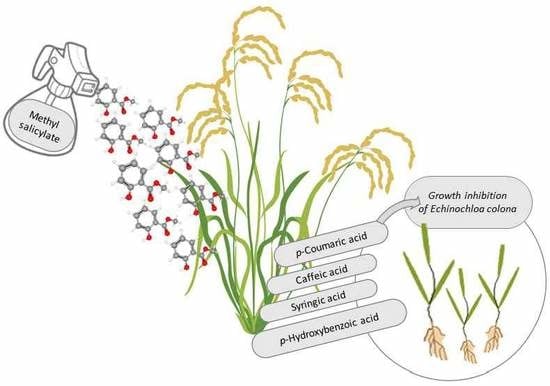
Elicitation of the Allelopathic Potential of Rice by Methyl Salicylate Treatment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Seeds
2.3. Experimental Design
2.4. Methyl Salicylate Treatment
2.5. Morphological Trait Measurements
2.6. Aqueous Extraction
2.7. Phytotoxic Activity
2.8. Determination of the Phenolic Acid Content
2.9. Statistical Analysis
3. Results and Discussion
3.1. Morphological Traits
3.2. Weed Population
3.3. Methyl Salicylate Treatments
3.4. Phenolic Acids
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cheng, F.; Cheng, Z. Research progress on the use of plant allelopathy in agriculture and the physiological and ecological mechanisms of allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef] [PubMed]
- Patni, B.; Chandra, H.; Mishra, A.P.; Guru, S.K.; Vitalini, S.; Iriti, M. Rice allelopathy in weed management–An integrated approach. Cell. Mol. Biol. 2018, 64, 84–93. [Google Scholar] [CrossRef] [PubMed]
- He, H.Q.; Shen, L.H.; Xiong, J.; Jia, X.L.; Lin, W.X.; Wu, H. Conditional genetic effect of allelopathy in rice (Oryza sativa L.) under different environmental conditions. Plant Growth Regul. 2004, 44, 211–218. [Google Scholar] [CrossRef]
- Parvez, S.S.; Parvez, M.M.; Fujii, Y.; Gemma, H. Allelopathic competence of Tamarindus indica L. root involved in plant growth regulation. Plant Growth Regul. 2003, 41, 39–148. [Google Scholar]
- Dilday, R.H.; Nastasi, P.; Smith, J.R. Allelopathic observations in rice (Oryza sativa L.) to ducksalad (Heteranthera limosa (SW) Willd). Proc. Arkansas Acad. Sci. 1989, 43, 21–22. [Google Scholar]
- Fujii, Y. The potential for biological control of paddy and aquatic weeds with allelopathy: allelopathic effect of some rice varieties. In Proceedings of the International Symposium on Biological Control and Integrated Management of Paddy and Aquatic Weeds, Tsukuba, Japan, 19–25 October 1992; pp. 305–320. [Google Scholar]
- Garrity, D.P.; Movillon, M.; Moddy, K. Differential weed suppression ability in upland rice cultivars. Agron. J. 1992, 84, 586–591. [Google Scholar] [CrossRef]
- Dilday, R.H.; Lin, J.; Yan, W.G. Identification of allelopathy in the USDA-ARS rice germplasm collection. Aust. J. Exp. Agric. 1994, 34, 901–910. [Google Scholar] [CrossRef]
- Olofsdotter, M.; Navarez, D.; Moody, K. Allelopathic potential in rice (Oryza sativa L.) germplasm. Ann. Appl. Biol. 1995, 127, 543–560. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, K.H.; Ahn, J.K.; Ju, H.J. Allelopathic potential evaluation of rice cultivars on Echinochloa crus galli. Korean J. Weed Sci. 1997, 17, 52–58. [Google Scholar]
- Chou, C.H. Roles of allelopathy in plant biodiversity and sustainable agriculture. Crit. Rev. Plant Sci. 1999, 18, 609–636. [Google Scholar] [CrossRef]
- Chung, I.M.; Ahn, J.K.; Kim, J.T.; Kim, C.S. Assessment of allelopathic potentiality and identification of allelopathic compounds on Korean local rice varieties. Korean J. Crop. Sci. 2000, 45, 44–49. [Google Scholar]
- Ahn, J.K.; Chung, I.M. Allelopathic potential of rice hulls on germination and seedling growth of barnyardgrass. Agron. J. 2000, 92, 1162–1167. [Google Scholar] [CrossRef]
- Chung, I.M.; Ahn, J.K.; Yun, S.J. Identification of allelopathic compounds from rice (Oryza sativa L.) straw and their biological activity. Can. J. Plant Sci. 2001, 81, 815–819. [Google Scholar] [CrossRef]
- Chung, I.M.; Ahn, J.K.; Yun, S.J. Assessment of allelopathic potential of barnyardgrass (Echinochloa crus-galli) on rice (Oryza sativa L.) cultivars. Crop Prot. 2001, 20, 921–928. [Google Scholar] [CrossRef]
- Ahn, J.K.; Hahn, S.J.; Kim, J.T.; Khanh, T.D.; Chung, I.M. Evaluation of allelopathic potential among rice (Oryza sativa L.) germplasm for control of Echinochloa crus-galli P. Beauv in the field. Crop Prot. 2005, 24, 413–419. [Google Scholar] [CrossRef]
- Jensen, L.B.; Courtis, B.; Shen, L.S.; Li, Z.K.; Olofsdotter, M.; Mauleon, R.P. Locating genes controlling allelopathic effects against barnyardgrass in upland rice. Agron. J. 2001, 93, 21–26. [Google Scholar] [CrossRef]
- Parvez, S.S.; Parvez, M.M.; Nishihara, E.; Gemma, H.; Fujii, Y. Tamarindus indica L. leaf is a source of allelopathic substance. Plant Growth Regul. 2003, 40, 107–115. [Google Scholar] [CrossRef]
- Khan, A.H.; Vaishya, R.D. Allelopathic effects of different crop residues on germination and growth of weeds. In Proceedings of the First National Symposium on Allelopathy in Agroecosystem (Agriculture & Forestry), CCS Haryana Agricultural University, Hisar, India, 12–14 February 1992; Tauro, P., Narwal, S.S., Eds.; Indian Society of Allelopathy: Hisar, India, 1992; pp. 59–60. [Google Scholar]
- Jafari, L.; Ghadiri, H.; Moradshahi, A. Allelopathic potential of rice (Oryza sativa L.) cultivars on barnyard grass (Echinochloa crus-galli). J. Agric. Sci. Technol. B 2011, 1, 853–864. [Google Scholar]
- Ashry, M.A.; Zein, A.A.; El-Nady, M.F.; Abdel-Dayem, Sh.M. Effect of potential allelopathic Egyptian rice cultivars against Echinochloa crus-galli and Echinochloa colona. J. Plant Prot. Path. 2012, 3, 629–644. [Google Scholar]
- Kaloumenos, N.S.; Chatzilazaridou, S.L.; Mylona, P.V.; Polidorosd, A.N.; Eleftherohorinos, I.G. Target-site mutation associated with cross-resistance to ALS-inhibiting herbicides in late watergrass (Echinochloa oryzicola Vasing.). Pest Manag. Sci. 2013, 69, 865–873. [Google Scholar] [CrossRef]
- Wilson, M.J.; Norswrthy, J.K.; Scott, R.C.; Gbur, E.E. Program approaches to control herbicide resistant barnyardgrass (Echinochloa crus-galli) in Midsouthern United States rice. Weed Technol. 2014, 25, 39–46. [Google Scholar] [CrossRef]
- Bajwa, A.A.; Jabran, K.; Shahid, M.; Ali, H.H.; Chauhan, B.S.; Ehsanullah. Eco-biology and management of Echinochloa crus-galli. Crop Prot. 2015, 75, 151–162. [Google Scholar] [CrossRef]
- Alam, M.A.; Hakim, M.A.; Juraimi, A.S.; Rafii, M.Y.; Hasan, M.M.; Aslani, F. Potential allelopathic effects of rice plant aqueous extracts on germination and seedling growth of some rice field common weeds. Ital. J. Agron. 2018, 13, 1066. [Google Scholar] [CrossRef]
- Bi, H.H.; Zeng, R.S.; Su, L.M.; An, M.; Luo, S.M. Rice allelopathy induced by methyl jasmonate and methyl salicylate. J. Chem. Ecol. 2007, 33, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Greenberg, J.T.; Holuigue, L. Editorial: Salicylic acid signaling networks. Front. Plant Sci. 2016, 7, 238. [Google Scholar] [CrossRef]
- Turner, J.G.; Ellis, C.; Evoto, A. The jasmonate signal pathway. Plant Cell 2002, 14, 153–164. [Google Scholar] [CrossRef]
- Lu, H. Dissection of salicylic acid-mediated defense signaling networks. Plant Signal. Behav. 2009, 4, 713–717. [Google Scholar] [CrossRef]
- An, Y.; Shen, Y.B.; Wu, L.J.; Zhang, Z.X. A change of phenolic acids content in poplar leaves induced by methyl salicylate and methyl jasmonate. J. For. Res. 2006, 17, 107–110. [Google Scholar] [CrossRef]
- Green, T.; Rogers, S.; Franzen, A.; Gentry, R.A. critical review of the literature to conduct a toxicity assessment for oral exposure to methyl salicylate. Crit. Rev. Toxicol. 2017, 47, 98–120. [Google Scholar] [CrossRef]
- Patni, B.; Guru, S.K. Morphophysiological and biochemical parameters associated with competitive ability of rice genotypes against weeds. Indian J. Plant Physiol. 2012, 17, 215–223. [Google Scholar]
- Johnson, D.E.; Dingkuhn, M.; Jones, M.P.; Mahamane, M.C. The influence of rice plant type on the effect of weed competition on Oryza sativa and Oryza glaberrima. Weed Res. 1998, 38, 207–216. [Google Scholar] [CrossRef]
- Mohammadi, G.R. Alternative weed control methods: A review. In Weed and Pest Control—Conventional and New Challenges, 1st ed.; Soloneski, S., Larramendy, M., Eds.; InTech: London, UK, 2013; pp. 117–159. [Google Scholar]
- Dass, A.; Shekhawat, K.; Choudhary, A.K.; Sepat, S.; Singh Rathore, S.; Mahajan, G.; Singh Chauhan, B. Weed management in rice using crop competition—A review. Crop Prot. 2017, 95, 45–52. [Google Scholar] [CrossRef]
- Chung, I.M.; Kim, K.H.; Ahn, J.K.; Lee, S.B.; Kim, S.H.; Hahn, S.J. Comparison of allelopathic potential of rice leaves, straw, and hull extracts on barnyardgrass. Agron J. 2003, 95, 1063–1070. [Google Scholar] [CrossRef]
- Jung, W.S.; Kim, K.H.; Ahn, J.K.; Hahn, S.J.; Chung, I.M. Allelopathic potential of rice (Oryza sativa L.) residues against Echinochloa crus galli. Crop Prot. 2004, 23, 211–218. [Google Scholar] [CrossRef]
- Awan, T.H.; Lim, C.A.A.; Ahmed, S.; Safdar, M.E.; Chauhan, B.S. Weed-competitive ability of a hybrid and an inbred rice cultivar in managing Ischaemum rugosum in fry-seeded rice. Pak. J. Agric. Sci. 2018, 55, 739–748. [Google Scholar]
- Wang, R.L.; Liu, S.W.; Xin, X.W.; Chen, S.; Peng, G.X.; Su, Y.J.; Song, Z.K. Phenolic acids contents and allelopathic potential of 10-cultivars of alfalfa and their bioactivity. Allelopath. J. 2017, 40, 63–70. [Google Scholar] [CrossRef]
- Li, Z.H.; Wang, Q.; Ruan, X.; Pan, C.D.; Jiang, D.A. Phenolics and plant allelopathy. Molecules 2010, 15, 8933–8952. [Google Scholar] [CrossRef]
- Bravo, H.R.; Copaja, S.V.; Lamborot, M. Phytotoxicity of phenolic acids from cereals. In Weed and Pest Control—Conventional and New Challenges, 1st ed.; Soloneski, S., Larramendy, M., Eds.; InTech: London, UK, 2013; pp. 37–49. [Google Scholar]
- Iriti, M.; Faoro, F. Chemical diversity and defence metabolism: how plants cope with pathogens and ozone pollution. Int. J. Mol. Sci. 2009, 10, 3371–3399. [Google Scholar] [CrossRef] [Green Version]


| Genotypes | Plant Height (cm) | Leaf No. | Leaf Area (cm2) | Dry Matter (g m−1) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Flowering | Maturity | Flowering | Maturity | Flowering | Maturity | Flowering | Maturity | |||||||||
| WF | W | WF | W | WF | W | WF | W | WF | W | WF | W | WF | W | WF | W | |
| PD-16 | 100.8 | 91.3 | 102.0 | 101.5 | 32.3 | 31.8 | 15.8 | 17.3 | 1442.3 | 1372.3 | 324.2 | 339.5 | 15.6 | 14.6 | 25.1 | 20.8 |
| UPR2916-211 | 93.3 | 84.8 | 104.8 | 100.6 | 29.1 | 32.2 | 22.1 | 20.0 | 1505.0 | 1466.6 | 649.6 | 524.3 | 19.8 | 17.1 | 25.5 | 22.3 |
| PSD-3 | 103.8 | 93.1 | 105.6 | 102.0 | 34.6 | 39.8 | 27.3 | 22.1 | 1784.3 | 1574.3 | 69.3 | 84.6 | 15.9 | 13.6 | 22.0 | 19.2 |
| UPR-2919-14-1-1 | 92.8 | 84.0 | 101.3 | 94.60 | 39.0 | 44.8 | 24.3 | 18.3 | 1900.6 | 1669.6 | 326.0 | 72.3 | 19.3 | 17.4 | 29.8 | 27.6 |
| UPR-2962-6-2-1 | 88.8 | 88.3 | 102.3 | 101.6 | 32.3 | 42.6 | 22.0 | 22.3 | 1151.6 | 1087.3 | 889.6 | 840.3 | 18.1 | 16.9 | 33.6 | 31.1 |
| UPR-2992-17-3-1 | 94.0 | 91.3 | 103.1 | 102.1 | 35.1 | 49.5 | 23.1 | 16.6 | 1497.6 | 956.0 | 39.3 | 70.6 | 18.5 | 15.3 | 23.3 | 18.7 |
| UPRI-2005-15 | 101.5 | 84.6 | 105.3 | 102.3 | 36.0 | 37.0 | 19.0 | 16.6 | 2002.3 | 1896.3 | 265.3 | 169.6 | 13.7 | 10.6 | 25.6 | 26.6 |
| UPR-2805-14-12 | 77.5 | 80.3 | 93.6 | 86.1 | 35.6 | 36.0 | 24.8 | 21.5 | 1380.0 | 823.6 | 417.3 | 387.6 | 8.7 | 6.9 | 24.8 | 21.2 |
| V3R11 | 89.8 | 82.1 | 99.8 | 98.3 | 46.8 | 49.1 | 27.8 | 26.0 | 1131.0 | 1107.3 | 681.3 | 814.0 | 16.9 | 15.3 | 26.1 | 22.5 |
| Govind | 82.5 | 79.1 | 84.6 | 83.3 | 36.3 | 38.8 | 21.5 | 21.3 | 984.3 | 949.3 | 157.3 | 326.0 | 18.2 | 17.4 | 28.4 | 26.7 |
| SEM ± | 0.63 | 0.72 | 0.45 | 0.75 | 0.13 | 0.38 | 0.17 | 0.30 | ||||||||
| LSD (p ≤ 0.05) | 3.74 | 4.31 | 26.94 | 4.49 | 82.29 | 22.91 | 1.04 | 1.79 | ||||||||
| Genotypes (G) | 6.06 | 3.00 | 6.42 | 4.51 | 97.64 | 55.73 | 1.99 | 6.90 | ||||||||
| Weed (W) | 8.57 | 5.69 | 9.08 | 6.38 | 138.09 | 78.82 | 2.81 | 9.76 | ||||||||
| G × W | 10.12 | 9.11 | 56.33 | 9.69 | 188.86 | 83.06 | 3.15 | 9.68 | ||||||||
| Genotypes | MeSA (mM) | E. colona Germination (%) | E. colona Root Length (cm) | E. colona Shoot Length (cm) | |||
|---|---|---|---|---|---|---|---|
| UPR-2992-17-3-1 | Shoot extract | Root extract | Shoot extract | Root extract | Shoot extract | Root extract | |
| Control | 86.3 | 77.7 | 6.8 | 6.8 | 5.8 | 5.9 | |
| 1 | 77.3 | 70.0 | 6.1 | 6.4 | 5.1 | 5.5 | |
| 2 | 69.7 | 64.7 | 5.5 | 5.9 | 4.5 | 5.3 | |
| 3 | 63.7 | 53.0 | 5.2 | 5.4 | 4.2 | 5.1 | |
| UPR-2962-6-2-1 | Control | 65.0 | 61.0 | 6.4 | 6.7 | 5.7 | 5.8 |
| 1 | 59.0 | 54.0 | 5.5 | 5.5 | 5.0 | 5.1 | |
| 2 | 46.7 | 42.3 | 4.9 | 4.7 | 4.0 | 4.6 | |
| 3 | 38.0 | 35.7 | 4.1 | 4.2 | 3.4 | 3.5 | |
| Govind | Control | 58.3 | 57.7 | 5.9 | 5.9 | 5.5 | 5.8 |
| 1 | 50.7 | 48.3 | 5.1 | 5.4 | 4.8 | 5.3 | |
| 2 | 43.3 | 39.0 | 4.4 | 4.7 | 4.1 | 4.6 | |
| 3 | 32.3 | 30.0 | 4.0 | 4.2 | 3.3 | 3.6 | |
| LSD (p ≤ 0.05) | 4.04 | 2.83 | 0.80 | 0.81 | 0.80 | 0.64 | |
| Genotypes (G) | 5.50 | 3.84 | 1.08 | 1.11 | 1.09 | 0.87 | |
| Methyl salicylate (MeSA) | 6.35 | 4.44 | 1.25 | 1.28 | 1.26 | 1.00 | |
| G × MeSA | 11.0 | 7.69 | 2.17 | 2.22 | 2.18 | 1.74 | |
| Phenolic Acid | Retention Time (min) |
|---|---|
| Syringic acid (S) | 9.9 |
| Vanillic acid (V) | 7.1 |
| p-Hydroxybenzoic acid (PHB) | 9.4 |
| p-Coumaric acid (PC) | 13.0 |
| Caffeic acid (CAF) | 11.6 |
| Protocatechuic acid PRO) | 5.57 |
| 8-Hydroxyquinoline (HQ) | 4.8 |
| Gallic acid (GAL) | 3.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patni, B.; Guru, S.K.; Iriti, M.; Vitalini, S. Elicitation of the Allelopathic Potential of Rice by Methyl Salicylate Treatment. Appl. Sci. 2019, 9, 4881. https://doi.org/10.3390/app9224881
Patni B, Guru SK, Iriti M, Vitalini S. Elicitation of the Allelopathic Potential of Rice by Methyl Salicylate Treatment. Applied Sciences. 2019; 9(22):4881. https://doi.org/10.3390/app9224881
Chicago/Turabian StylePatni, Babita, Sudhir Kumar Guru, Marcello Iriti, and Sara Vitalini. 2019. "Elicitation of the Allelopathic Potential of Rice by Methyl Salicylate Treatment" Applied Sciences 9, no. 22: 4881. https://doi.org/10.3390/app9224881
APA StylePatni, B., Guru, S. K., Iriti, M., & Vitalini, S. (2019). Elicitation of the Allelopathic Potential of Rice by Methyl Salicylate Treatment. Applied Sciences, 9(22), 4881. https://doi.org/10.3390/app9224881