Architecture and Viability of the Biofilms Formed by Nine Listeria Strains on Various Hydrophobic and Hydrophilic Materials
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culturing Conditions
2.2. Biofilm Formation
2.3. Determination of Cell Surface Hydrophobicity
2.4. Statistical Analysis
3. Results
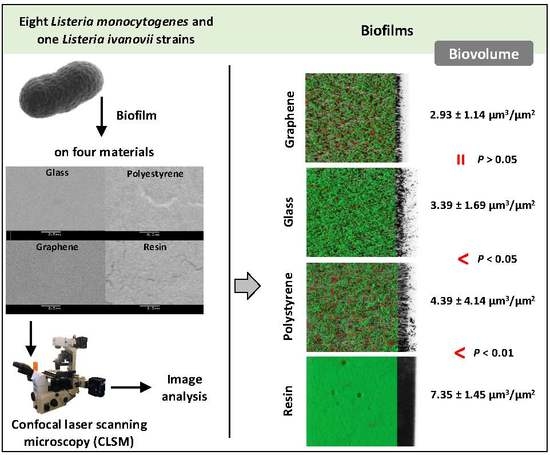
3.1. Biofilm Determination
3.2. Cell Surface Hydrophobicity
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rothrock, M.J.; Micciche, A.C.; Bodie, A.R.; Ricke, S.C. Listeria occurrence and potential control strategies in alternative and conventional poultry processing and retail. Front. Sust. Food Syst. 2019, 3, 33. [Google Scholar] [CrossRef]
- Davis, M.L.; Ricke, S.C.; Donalson, J.R. Establishment of Listeria monocytogenes in the gastrointestinal tract. Microorganisms 2019, 7, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanciotti, R.; Braschi, G.; Patrignani, F.; Gobbetti, M.; De Angelis, M. How Listeria monocytogenes shapes its proteome in response to natural antimicrobial compounds. Front. Microbiol. 2019, 10, 437. [Google Scholar] [CrossRef] [PubMed]
- Doijad, S.P.; Barbuddhe, S.B.; Garg, S.; Poharkar, K.V.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Chakraborty, T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE 2015, 10, e0137046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EFSA; ECDC. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- CDC. Listeria (listeriosis). Centers for Disease Control and Prevention. Available online: https://www.cdc.gov/listeria/index.html (accessed on 14 September 2019).
- Pohl, A.M.; Pouillot, R.; Van Doren, J.M. Changing US population demographics: What does this mean for listeriosis incidence and exposure? Foodborne Pathog. Dis. 2017, 14, 524–530. [Google Scholar] [CrossRef] [Green Version]
- Leclercq, A.; Moura, A.; Vales, G.; Tessaud-Rita, N.; Aguilhon, C.; Lecuit, M. Listeria thailandensis sp. nov. Int. J. Syst. Microbiol. 2019, 69, 74–81. [Google Scholar] [CrossRef]
- Wagner, M.; McLauchlin, J. Biology. In Handbook of Listeria Monocytogenes; Dongyou, L., Ed.; CRC Press: Boca Raton, FL, USA, 2008; pp. 3–25. [Google Scholar]
- Orsi, R.H.; Wiedmann, M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016, 100, 5273–5287. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Quingping, W.; Jumei, Z.; Moutong, C.; Zéan, Y. Prevalence, antibiotic resistance and genetic diversity of Listeria monocytogenes isolated from retail ready-to-eat foods in China. Food Control 2015, 47, 340–347. [Google Scholar] [CrossRef]
- Capita, R.; Felices-Mercado, A.; García-Fernández, C.; Alonso-Calleja, C. Characterization of Listeria monocytogenes originating from the Spanish meat-processing chain. Foods 2019, 8, 542. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Vázquez-Sánchez, D.; López-Cabo, M. Current knowledge on Listeria monocytogenes biofilms in food-related environments: Incidence, resistance to biocides, ecology and biocontrol. Foods 2018, 7, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Rivas, F.; Ripolles-Avila, C.; Fontecha-Umaña, F.; Ríos-Castillo, A.G.; Rodríguez-Jerez, J.J. Biofilms in the spotlight: Detection, quantification and elimination at a food industry level. Comp. Rev. Food Sci. Food Saf. 2018, 17, 1261–1276. [Google Scholar] [CrossRef] [Green Version]
- Piercey, M.J.; Ells, T.C.; Macintosh, A.J.; Hansen, L.T. Variations in biofilm formation, desiccation resistance and benzalkonium chloride susceptibility among Listeria monocytogenes strains isolated in Canada. Int. J. Food Microbiol. 2017, 257, 254–261. [Google Scholar] [CrossRef] [Green Version]
- González-Machado, C.; Capita, R.; Riesco-Peláez, F.; Alonso-Calleja, C. Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development. PLoS ONE 2018, 13, e0200011. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Peng, L.-H.; Zhang, S.; Zhou, S.; Yoshida, A.; Osatomi, K.; Bellou, N.; Guo, X.-P.; Dobretsov, S.; Yang, J.-L. Polyurethane, epoxy resin and polydimethylsiloxane altered biofilm formation and mussel settlement. Chemosphere 2019, 218, 599–608. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; Carballo, J.; García-Fernández, C.; Capita, R.; Alonso-Calleja, C. Structure and viability of 24- and 72-h-old biofilms formed by four pathogenic bacteria on polystyrene and glass contact surfaces. Food Microbiol. 2018, 76, 513–517. [Google Scholar] [CrossRef]
- Ben Abdallah, F.; Lagha, R.; Said, K.; Kallel, H.; Gharbi, J. Detection of cell surface hydrophobicity, biofilm and fimbriae genes in Salmonella isolated from Tunisian clinical and poultry meat. Iran J. Public Health 2014, 43, 423–431. [Google Scholar]
- Yilbas, B.S.; Ibrahim, A.; Ali, H.; Khaled, M.; Laoui, T. Hydrophobic and optical characteristics of graphene and graphene oxide films transferred onto functionalized silica particles deposited glass surface. Appl. Surf. Sci. 2018, 442, 213–223. [Google Scholar] [CrossRef]
- Jin, F.-L.; Li, X.; Park, S.-J. Synthesis and application of epoxy resins: A review. J. Ind. Eng. Chem. 2015, 29, 1–11. [Google Scholar] [CrossRef]
- Kurantowicz, N.; Sawosz, E.; Jaworski, S.; Kutwin, M.; Strojny, B.; Wierzbicki, M.; Szeliga, J.; Hotowy, A.; Lipińska, L.; Koziński, R.; et al. Interaction of graphene family materials with Listeria monocytogenes and Salmonella enterica. Nanoscale Res. Lett. 2015, 10, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szunerits, S.; Boukherroub, R. Antibacterial activity of graphene-based materials. J. Mater. Chem. B 2016, 4, 6892–6912. [Google Scholar] [CrossRef] [Green Version]
- Dybowska-Sarapuk, Ł.; Kotela, A.; Krzemiński, J.; Wróblewska, M.; Marchel, H.; Romaniec, M.; Łęgosz, P.; Jakubowska, M. Graphene nanolayers as a new method for bacterial biofilm prevention: Preliminary results. J. AOAC Int. 2017, 100, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Choi, N.-Y.; Bae, Y.M.; Lee, S.-Y. Cell surface properties and biofilm formation of pathogenic bacteria. Food Sci. Biotechnol. 2015, 24, 2257–2264. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Bruini, I.; Di Ciccio, P.A.; Zanardi, E.; Ghidini, S.; Ianieri, A. Listeria monocytogenes biofilms in the wonderland of Food Industry. Pathogens 2017, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, H.; Suda, T.; Tanaka, Y.; Kimura, B. Cellular hydrophobicity of Listeria monocytogenes involves initial attachment and biofilm formation on the surface of polyvinyl chloride. Lett. Appl. Microbiol. 2010, 50, 618–625. [Google Scholar] [CrossRef]
- Lee, B.-H.; Hébraud, M.; Bernardi, T. Increased adhesion of Listeria monocytogenes strains to abiotic surfaces under cold stress. Front. Microbiol. 2017, 8, 2221. [Google Scholar] [CrossRef]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef] [Green Version]
- Ripolles-Avila, C.; Hascoët, A.S.; Guerrero-Navarro, A.E.; Rodríguez-Jerez, J.J. Establishment of incubation conditions to optimize the in vitro formation of mature Listeria monocytogenes biofilms on food-contact surfaces. Food Control 2018, 92, 240–248. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Capita, R.; García-Fernández, C.; Alonso-Calleja, C. Effects of Bacteriophage P100 at different concentrations on the structural parameters of Listeria monocytogenes biofilms. J. Food Prot. 2018, 81, 2040–2044. [Google Scholar] [CrossRef]
- Ripolles-Avila, C.; Cervantes-Huaman, B.H.; Hascoët, A.S.; Yuste, J.; Rodríguez-Jerez, J.J. Quantification of mature Listeria monocytogenes biofilms: A comparison of different methods. Int. J. Food Microbiol. 2019, 289, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Melcón, C.; Capita, R.; Rodríguez-Jerez, J.J.; Martínez-Suárez, J.V.; Alonso-Calleja, C. Effect of low doses of disinfectants on the biofilm-forming ability of Listeria monocytogenes. Foodborne Pathog. Dis. 2019, 16, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Melcón, C.; Riesco-Peláez, F.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Susceptibility of Listeria monocytogenes planktonic cultures and biofilms to sodium hypochlorite and benzalkonium chloride. Food Microbiol. 2019, 82, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Norwood, D.E.; Gilmour, A. Adherence of Listeria monocytogenes strains to stainless steel coupons. J. Appl. Microbiol. 1999, 86, 576–582. [Google Scholar] [CrossRef]
- Marsh, E.J.; Luo, H.; Wang, H. A three-tiered approach to differentiate Listeria monocytogenes biofilm-forming abilities. FEMS Microbiol. Lett. 2003, 228, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Mosquera-Fernández, M.; Rodríguez-López, P.; Cabo, M.L.; Balsa-Canto, E. Numerical spatio-temporal characterization of Listeria monocytogenes biofilms. Int. J. Food Microbiol. 2014, 182, 26–36. [Google Scholar] [CrossRef] [Green Version]
- Guilbaud, M.; Piveteau, P.; Desvaux, M.; Brisse, S.; Briandet, R. Exploring the diversity of Listeria monocytogenes biofilm architecture by high-throughput confocal laser scanning microscopy and the predominance of the honeycomb-like morphotype. Appl. Environ. Microbiol. 2015, 81, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
- Reis-Teixeira, F.B.D.; Alves, V.F.; de Martinis, E.C.P. Growth, viability and architecture of biofilms of Listeria monocytogenes formed on abiotic surfaces. Braz. J. Microbiol. 2017, 48, 587–591. [Google Scholar] [CrossRef]
- Pang, X.; Wong, C.; Chung, H.-J.; Yuk, H.-G. Biofilm formation of Listeria monocytogenes and its resistance to quaternary ammonium compounds in a simulated salmon processing environment. Food Control 2019, 98, 200–208. [Google Scholar] [CrossRef]
- Alonso-Calleja, C.; Gómez-Fernández, S.; Carballo, J.; Capita, R. Prevalence, molecular typing and determination of the biofilm-forming ability of Listeria monocytogenes serotypes from poultry meat and poultry preparations in Spain. Microorganisms 2019, 7, 529. [Google Scholar] [CrossRef] [Green Version]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernandes, A.; Araújo, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control 2014, 35, 386–391. [Google Scholar] [CrossRef]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Moreno, A.M.; González-Martín, M.L.; Pérez-Giraldo, C.; Bruque, J.M.; Gómez-García, A.C. Serum as a factor influencing adhesion of Enterococcus faecalis to glass and silicone. Appl. Environ. Microbiol. 2002, 68, 5784–5787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallardo-Moreno, A.M.; González-Martín, M.L.; Pérez-Giraldo, C.; Garduno, E.; Bruque, J.M.; Gómez-García, A.C. Thermodynamic analysis of growth temperature dependence in the adhesion of Candida parapsilosis to polystyrene. Appl. Environ. Microbiol. 2002, 68, 2610–2613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norouzi, F.; Mansouri, S.; Moradi, M.; Razavi, M. Comparison of cell surface hydrophobicity and biofilm formation among ESBL- and non-ESBL-producing Pseudomonas aeruginosa clinical isolates. Afr. J. Microbiol. Res. 2010, 4, 1143–1147. [Google Scholar]
- Patel, J.; Sharma, M.; Ravishakar, S. Effect of curli expression and hydrophobicity of Escherichia coli O157:H7 on attachment to fresh produce surfaces. J. Appl. Microbiol. 2011, 110, 737–745. [Google Scholar] [CrossRef]
- Dourou, D.; Beauchamp, C.S.; Yoon, Y.; Geornaras, I.; Belk, K.E.; Smith, G.C.; Nychas, G.-J.E.; Sofos, J.N. Attachment and biofilm formation by Escherichia coli O157:H7 at different temperatures, on various food-contact surfaces encountered in beef processing. Int. J. Food Microbiol. 2011, 149, 262–268. [Google Scholar] [CrossRef]
- Chavant, P.; Martinie, B.; Meylheuc, T.; Bellon-Fontaine, M.N.; Hebraud, M. Listeria monocytogenes LO28: Surface physicochemical properties and ability to form biofilms at different temperatures and growth phases. Appl. Environ. Microbiol. 2002, 68, 728–737. [Google Scholar] [CrossRef] [Green Version]
- Piercey, M.J.; Hingston, P.A.; Hansen, T.L. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- Tomičić, R.M.; Čabarkapa, I.S.; Vukmirović, Ð.M.; Lević, J.D.; Tomičić, Z.M. Influence of growth conditions on biofilm formation of Listeria monocytogenes. Food Feed Res. 2016, 43, 19–24. [Google Scholar] [CrossRef]
- Dong, H.S.; Qi, S.J. Realising the potential of graphene-based materials for biosurfaces—A future perspective. Biosurf. Biotribol. 2015, 1, 229–248. [Google Scholar] [CrossRef] [Green Version]



| Dye | ||
|---|---|---|
| Parameter | SYTO 9 | Propidium Iodide |
| Pinhole | 44 µm | 44 µm |
| Laser Wavelength | 488 nm: 0.20% | 561 nm: 0.20% |
| Excitation Wavelength | 483 nm | 305 nm |
| Emission Wavelength | 500 nm | 617 nm |
| Detection Wavelength | 450–560 nm | 560–700 nm |
| Imaging Device | LSM 800/GaAsP-Pmt2 | LSM 800/Airyscan |
| Detector | GaAsP | Airyscan |
| Detector Gain | 700 V | 700 V |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Architecture and Viability of the Biofilms Formed by Nine Listeria Strains on Various Hydrophobic and Hydrophilic Materials. Appl. Sci. 2019, 9, 5256. https://doi.org/10.3390/app9235256
Rodríguez-Melcón C, Alonso-Calleja C, Capita R. Architecture and Viability of the Biofilms Formed by Nine Listeria Strains on Various Hydrophobic and Hydrophilic Materials. Applied Sciences. 2019; 9(23):5256. https://doi.org/10.3390/app9235256
Chicago/Turabian StyleRodríguez-Melcón, Cristina, Carlos Alonso-Calleja, and Rosa Capita. 2019. "Architecture and Viability of the Biofilms Formed by Nine Listeria Strains on Various Hydrophobic and Hydrophilic Materials" Applied Sciences 9, no. 23: 5256. https://doi.org/10.3390/app9235256
APA StyleRodríguez-Melcón, C., Alonso-Calleja, C., & Capita, R. (2019). Architecture and Viability of the Biofilms Formed by Nine Listeria Strains on Various Hydrophobic and Hydrophilic Materials. Applied Sciences, 9(23), 5256. https://doi.org/10.3390/app9235256