OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera
Abstract
:1. Introduction
2. Materials and Methods
2.1. F98 Rat Glioma Model
2.2. OKN-007 Treatment
2.3. Blood Collection and Serum Separation from the Whole Blood
2.4. ESI-MS
2.5. LOOCV Mass Peak Analysis and Statistics
2.6. Tandem MS/MS and Bioinformatic Analysis
2.7. Test Metrics
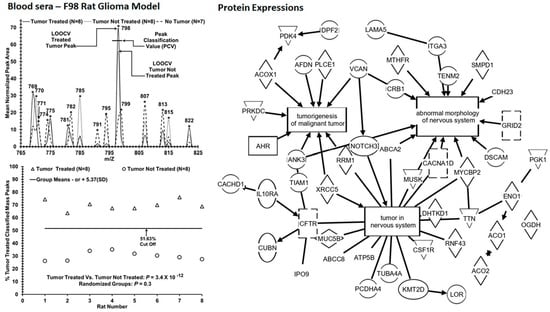
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lapointe, S.; Perry, A.; Butowski, N.A. Primary brain tumours in adults. Lancet 2018, 392, 432–446. [Google Scholar] [CrossRef]
- Garteiser, P.; Doblas, S.; Watanabe, Y.; Saunders, D.; Hoyle, J.; Lerner, M.; He, T.; Floyd, R.A.; Towner, R.A. Multiparametric assessment of the anti-glioma properties of OKN007 by magnetic resonance imaging. J. Magn. Reson. Imaging 2010, 31, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Towner, R.A.; Gillespie, D.L.; Schwager, A.; Saunders, D.G.; Smith, N.; Njoku, C.E.; Krysiak, R.S., 3rd; Larabee, C.; Iqbal, H.; Floyd, R.A.; et al. Regression of glioma tumor growth in F98 and U87 rat glioma models by the Nitrone OKN-007. Neuro Oncol. 2013, 15, 330–340. [Google Scholar] [CrossRef] [Green Version]
- Coutinho de Souza, P.; Smith, N.; Atolagbe, O.; Ziegler, J.; Njoku, C.; Lerner, M.; Ehrenshaft, M.; Mason, R.P.; Meek, B.; Plafker, S.M.; et al. OKN-007 decreases free radical levels in a preclinical F98 rat glioma model. Free Radic. Biol. Med. 2015, 87, 157–168. [Google Scholar] [CrossRef] [PubMed]
- De Souza, P.C.; Smith, N.; Pody, R.; He, T.; Njoku, C.; Silasi-Mansat, R.; Lupu, F.; Meek, B.; Chen, H.; Dong, Y.; et al. OKN-007 decreases VEGFR-2 levels in a preclinical GL261 mouse glioma model. Am. J. Nucl. Med. Mol. Imaging 2015, 5, 363–378. [Google Scholar]
- Towner, R.A.; Smith, N.; Saunders, D.; Brown, C.A.; Cai, X.; Ziegler, J.; Mallory, S.; Dozmorov, M.G.; Coutinho De Souza, P.; Wiley, G.; et al. OKN-007 Increases temozolomide (TMZ) Sensitivity and Suppresses TMZ-Resistant Glioblastoma (GBM) Tumor Growth. Transl. Oncol. 2019, 12, 320–335. [Google Scholar] [CrossRef] [PubMed]
- Coutinho de Souza, P.; Mallory, S.; Smith, N.; Saunders, D.; Li, X.N.; McNall-Knapp, R.Y.; Fung, K.M.; Towner, R.A. Inhibition of Pediatric Glioblastoma Tumor Growth by the Anti-Cancer Agent OKN-007 in Orthotopic Mouse Xenografts. PLoS ONE 2015, 10, e0134276. [Google Scholar]
- Thomas, L.; Smith, N.; Saunders, D.; Zalles, M.; Gulej, R.; Lerner, M.; Fung, K.-M.; Carcaboso, A.M.; Towner, R.A. OKlahoma Nitrone-007: Novel treatment for diffuse intrinsic pontine glioma. J. Transl. Med. 2020, 18, 424. [Google Scholar] [CrossRef]
- De Souza, P.C.; Balasubramanian, K.; Njoku, C.; Smith, N.; Gillespie, D.; Schwager, A.; Abdullah, O.; Ritchey, J.W.; Fung, K.-M.; Saunders, D.; et al. OKN-007 decreases tumor necrosis and tumor cell proliferation and increases apoptosis in a preclinical F98 rat glioma model. J. Magn. Reson. Imaging 2015, 42, 1582–1591. [Google Scholar] [CrossRef]
- Barth, R.F.; Kaur, B. Rat brain tumor models in experimental neuro-oncology: The C6, 9L, T9, RG2, F98, BT4C, RT-2 and CNS-1 gliomas. J. Neuro-Oncol. 2009, 94, 299–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, N.; Saunders, D.; Jensen, R.L.; Towner, R.A. Association of decreased levels of lipopolysaccharide-binding protein with OKN-007–induced regression of tumor growth in an F98 rat glioma model. J. Neurosurg. 2020, 133, 1695–1703. [Google Scholar] [CrossRef]
- Hocker, J.R.; Peyton, M.D.; Lerner, M.R.; Mitchell, S.L.; Lightfoot, S.A.; Lander, T.J.; Bates-Albers, L.M.; Vu, N.T.; Hanas, R.J.; Kupiec, T.C.; et al. Serum discrimination of early-stage lung cancer patients using electrospray-ionization mass spectrometry. Lung Cancer 2011, 74, 206–211. [Google Scholar] [CrossRef]
- Hocker, J.R.; Mohammed, A.; Aston, C.E.; Brewer, M.; Lightfoot, S.A.; Rao, C.V.; Hanas, J.S. Mass profiling of serum to distinguish mice with pancreatic cancer induced by a transgenic kras mutation. Int. J. Cancer 2013, 133, 2662–2671. [Google Scholar] [PubMed] [Green Version]
- Hocker, J.R.; Postier, R.G.; Li, M.; Lerner, M.R.; Lightfoot, S.A.; Peyton, M.D.; Deb, S.J.; Baker, C.M.; Williams, T.L.; Hanas, R.J.; et al. Discriminating patients with early-stage pancreatic cancer or chronic pancreatitis using serum electrospray mass profiling. Cancer Lett. 2015, 359, 314–324. [Google Scholar] [CrossRef] [Green Version]
- Dittmar, G.A.G.; Wilkinson, C.R.M.; Jedrzejewski, P.T.; Finley, D. Role of a Ubiquitin-Like Modification in Polarized Morphogenesis. Science 2002, 295, 2442–2446. [Google Scholar] [CrossRef] [PubMed]
- Altman, D.G.; Bland, J.M. Statistics Notes: Diagnostic tests 2: Predictive values. BMJ 1994, 309, 102. [Google Scholar] [CrossRef] [Green Version]
- Altman, D.G.; Bland, J.M. Diagnostic tests. 1: Sensitivity and specificity. BMJ 1994, 308, 1552. [Google Scholar] [CrossRef] [Green Version]
- Andreollo, N.A.; Santos, E.F.; Araújo, M.R.; Lopes, L.R. Rat’s age versus human’s age: What is the relationship? Arq. Bras. Cir. Dig. 2012, 25, 49–51. [Google Scholar] [CrossRef] [Green Version]
- Rat Behavior and Biology. Rat Health Notes. How Old Is a Rat In Human Years? Last Updated 29 November 2012. Available online: http://www.ratbehavior.org/RatYears (accessed on 8 September 2021).
- Quinn, R. Comparing rat’s to human’s age: How old is my rat in people years? Nutrition 2005, 21, 775–777. [Google Scholar] [CrossRef]
- Lyden, P.D.; Shuaib, A.; Lees, K.R.; Davalos, A.; Davis, S.M.; Diener, H.C.; Grotta, J.C.; Ashwood, T.J.; Hardemark, H.G.; Svensson, H.H.; et al. Safety and tolerability of NXY-059 for acute intracerebral hemorrhage: The CHANT Trial. Stroke 2007, 38, 2262–2269. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.J.; Rhoney, D.H. NXY-059: Review of Neuroprotective Potential for Acute Stroke. Ann. Pharmacother. 2006, 40, 461–471. [Google Scholar] [CrossRef] [Green Version]
- Towner, R.A.; Saunders, D.; Lerner, M.; Silasi Mansat, R.; Yuan, T.; Barber, D.; Faakye, J.; Nyul-Toth, A.; Csiszar, A.; Greenwood-Van Meerveld, B.; et al. Temporary opening of the blood-brain barrier with the nitrone compound OKN-007. Am. J. Nucl. Med. Mol. Imaging 2021, 11, 363–373. [Google Scholar]
- Gharibi, A.; La Kim, S.; Molnar, J.; Brambilla, D.; Adamian, Y.; Hoover, M.; Hong, J.; Laurelin, W.; Wolfenden, L.; Kelber, J.A. ITGA1 is a pre-malignant biomarker that promotes therapy resistance and metastatic potential in pancreatic cancer. Sci. Rep. 2017, 7, 10060. [Google Scholar] [CrossRef]
- Price, J.C.; Azizi, E.; Naiche, L.A.; Parvani, J.G.; Shukla, P.; Kim, S.; Slack-Davis, J.K.; Pe’Er, D.; Kitajewski, J.K. Notch3 signaling promotes tumor cell adhesion and progression in a murine epithelial ovarian cancer model. PLoS ONE 2020, 15, e0233962. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Moses-Gardner, A.; Huang, J.; Smith, E.R.; Moses, M.A. ITGA2 as a potential nanotherapeutic target for glioblastoma. Sci. Rep. 2019, 9, 6195. [Google Scholar] [CrossRef] [Green Version]
- Kovářová, D.; Plachý, J.; Kosla, J.; Trejbalová, K.; Čermák, V.; Hejnar, J. Downregulation of HOPX Controls Metastatic Behavior in Sarcoma Cells and Identifies Genes Associated with Metastasis. Mol. Cancer Res. 2013, 11, 1235–1247. [Google Scholar] [CrossRef] [Green Version]
- Jiang, C.; Zhou, Y.; Huang, Y.; Wang, Y.; Wang, W.; Kuai, X. Overexpression of ADAMTS-2 in tumor cells and stroma is predictive of poor clinical prognosis in gastric cancer. Hum. Pathol. 2019, 84, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Soichi, O.; Masanori, N.; Hideo, T.; Kazunori, A.; Nobuya, I.; Jun-ichi, K. Clinical significance of ABCA2’ a possible molecular marker for oligodendrogliomas. Neurosurgery 2007, 60, 707–714, discussion 714. [Google Scholar] [PubMed]
- Xu, G.; Li, J.Y. ATP5A1 and ATP5B are highly expressed in glioblastoma tumor cells and endothelial cells of microvascular proliferation. J. Neuro-Oncol. 2016, 126, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Fu, W.-M. Identification of differential splicing genes in gliomas using exon expression profiling. Mol. Med. Rep. 2015, 11, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Li, Y.; Liu, S.; Chen, Q.; Liu, Y. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. OncoTargets Ther. 2019, 12, 4141–4152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawataki, T.; Yamane, T.; Naganuma, H.; Rousselle, P.; Andurén, I.; Tryggvason, K.; Patarroyo, M. Laminin isoforms and their integrin receptors in glioma cell migration and invasiveness: Evidence for a role of alpha5-laminin(s) and alpha3beta1 integrin. Exp. Cell Res. 2007, 313, 3819–3831. [Google Scholar] [CrossRef] [PubMed]
- Dhar, S.S.; Zhao, D.; Lin, T.; Gu, B.; Pal, K.; Wu, S.; Alam, H.; Lv, J.; Yun, K.; Gopalakrishnan, V.; et al. MLL4 Is Required to Maintain Broad H3K4me3 Peaks and Super-Enhancers at Tumor Suppressor Genes. Mol. Cell 2018, 70, 825–841.e6. [Google Scholar] [CrossRef] [Green Version]
- Erdem-Eraslan, L.; Gao, Y.; Kloosterhof, N.K.; Atlasi, Y.; Demmers, J.; Sacchetti, A.; Kros, J.M.; Sillevis Smitt, P.; Aerts, J.; French, P.J. Mutation specific functions of EGFR result in a mutation-specific downstream pathway activation. Eur. J. Cancer 2015, 51, 893–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pierfelice, T.J.; Schreck, K.; Dang, L.; Asnaghi, L.; Gaiano, N.; Eberhart, C.G. Notch3 Activation Promotes Invasive Glioma Formation in a Tissue Site-Specific Manner. Cancer Res. 2011, 71, 1115–1125. [Google Scholar] [CrossRef] [Green Version]
- Arslan, F.; Bosserhoff, A.K.; Nickl-Jockschat, T.; Doerfelt, A.; Bogdahn, U.; Hau, P. The role of versican isoforms V0/V1 in glioma migration mediated by transforming growth factor-beta2. Br. J. Cancer 2007, 96, 1560–1568. [Google Scholar] [CrossRef]
- Zhou, K.; Liu, Y.; Zhao, Z.; Wang, Y.; Huang, L.; Chai, R.; Li, G.; Jiang, T. ABCC8 mRNA expression is an independent prognostic factor for glioma and can predict chemosensitivity. Sci. Rep. 2020, 10, 12682. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, Y.; Zheng, P. Downregulation of ADAMTS18 May Serve as a Poor Prognostic Biomarker for Cervical Cancer Patients. Appl. Immunohistochem. Mol. Morphol. 2018, 26, 670–675. [Google Scholar] [CrossRef]
- Jin, H.; Wang, X.; Ying, J.; Wong, A.H.Y.; Li, H.; Lee, K.Y.; Srivastava, G.; Chan, A.T.C.; Yeo, W.; Ma, B.; et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene 2007, 26, 7490–7498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.; Wang, W.; Cheng, H. Co-expression network analysis of gene expression profiles of HER2+ breast cancer-associated brain metastasis. Oncol. Lett. 2018, 16, 7008–7019. [Google Scholar] [CrossRef] [Green Version]
- Afghani, N.; Mehta, T.; Wang, J.; Tang, N.; Skalli, O.; Quick, Q.A. Microtubule actin cross-linking factor 1, a novel target in glioblastoma. Int. J. Oncol. 2017, 50, 310–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galatenko, V.V.; Maltseva, D.V.; Galatenko, A.V.; Rodin, S.; Tonevitsky, A.G. Cumulative prognostic power of laminin genes in colorectal cancer. BMC Med. Genom. 2018, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Gamble, J.T.; Reed-Harris, Y.; Barton, C.L.; La Du, J.; Tanguay, R.; Greenwood, J.A. Quantification of glioblastoma progression in zebrafish xenografts: Adhesion to laminin alpha 5 promotes glioblastoma microtumor formation and inhibits cell invasion. Biochem. Biophys. Res. Commun. 2018, 506, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Costanza, B.; Umelo, I.A.; Bellier, J.; Castronovo, V.; Turtoi, A. Stromal Modulators of TGF-β in Cancer. J. Clin. Med. 2017, 6, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalles, M.; Smith, N.; Saunders, D.; Lerner, M.; Fung, K.; Battiste, J.; Towner, R.A. A tale of two multi-focal therapies for glioblastoma: An antibody targeting ELTD1 and nitrone-based OKN-007. J. Cell. Mol. Med. 2021, 26, 520–582. [Google Scholar] [CrossRef]



| Test Metrics (Group 1 vs. Group 2, n Values) | Mean (SD) | Mean (SD) | True Positive Group 1 | False Positive Group 1 | True Negative Group 2 | False Negative Group 2 | p-Value True Pathology [Random Groups] |
|---|---|---|---|---|---|---|---|
| Tumor Treated (8) vs. Tumor Not Treated (8) [ROC area = 1] | 0.70 | 0.30 | 8/8 | 0/0 | 8/8 | 0/0 | 3.4 × 10−12 |
| (0.03) | (0.03) | (100%) | (0%) | (100%) | (0%) | [0.3] | |
| Tumor Treated (n = 8) vs. No Tumor (control: n = 7) [ROC area = 1] | 0.73 | 0.29 | 8/8 | 0/0 | 7/7 | 0/0 | 2.4 × 10−11 |
| (0.03) | (0.04) | (100%) | (0%) | (100%) | (0%) | [0.41] | |
| Tumor Not Treated (8) vs. No Tumor (control: n = 7) [ROC area = 1] | 0.73 | 0.29 | 8/8 | 0/0 | 7/7 | 0/0 | 2.5 × 10−11 |
| (0.03) | (0.04) | (100%) | (0%) | (100%) | (0%) | [0.41] |
| Decreased in Serum from Treated Brain Tumor Animals Relative to Non-Treated Animals | |||||
| Protein | Pathway (IPA) | Protein | Pathway (IPA) | Protein | Pathway (IPA) |
| ABCA2 | B, C, CN, G, MM, MN, T | DDX1 | MM | NAT6 | |
| ACO2 | C, N | DHTKD1 | B, C, CN, G | NOTCH3 | B, C, CM, CN, G, N |
| ADGRE1 | DPYSL3 | C, MN, N | NSUN6 | ||
| AHR | CM, MM, N | FH | CM, MM, N, T | PDE11A | C, |
| ATP5B | B, CN, G, T | INTS12 | PIGS | ||
| CACHD1 | IPO9 | T | SMG5 | T | |
| CES2 | ITGA3 | C, MM, MN, N, T | TMEM132D | C, | |
| CFTR | C, CN, G, MM, T | KMT2D | C, CM, CN, G | TTBK2 | N |
| CNTN2 | MN | LOC108348049 | TTN | B, C, CN, G, MM, N | |
| CRB1 | MN | LOC690425 | VCAN | C, CM, MM, MN, N | |
| CSF1R | C, CM, CN, G, MM, N | MYCBP2 | C, CN, G, MM, MN | XRCC5 | C, CN, MM, N |
| CUBN | C, T | MYH3 | ZBTB37 | ||
| Increased in Serum from Treated Brain Tumor Animals Relative to Non-Treated Animals | |||||
| Protein | Pathway (IPA) | Protein | Pathway (IPA) | Protein | Pathway (IPA) |
| ABCA6 | GRID2 | MM, MN, N, T | MUC16 | ||
| ABCC8 | CN, MM, N, T | HADHA | MM, N | MUC19 | |
| ACO1 | C, T | LAMA5 | C, CM, N | MUSK | B, CN, G, MM, MN, N |
| ACOX1 | C | LOC102554371 | PAFAH2 | N | |
| ADAMTS18 | C, | LOC103691264 | PCDHA4 | B, C, CN, G | |
| ADGRF5 | C, CM, MM | LOC685544 | PCYT1A | MM, T | |
| AMACR | MM, N | LOR | C, | PLCE1 | C, N |
| CACNA1D | C, CM, CN, MN, T | MACF1 | MM | PTPN22 | N |
| CRYBG3 | MCM3AP | MM, T | REV3L | MM, N | |
| DHRSX | C, | MFSD12 | TBC1D23 | ||
| DSCAM | C, MM, MN | MICAL2 | C, | TENM2 | C, MN |
| EEF1AKMT1 | MRVI1 | MM | TUBA4A | ||
| EXNEF | C, | MTTP | VWA8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Towner, R.A.; Hocker, J.; Smith, N.; Saunders, D.; Battiste, J.; Hanas, J. OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera. Brain Sci. 2022, 12, 100. https://doi.org/10.3390/brainsci12010100
Towner RA, Hocker J, Smith N, Saunders D, Battiste J, Hanas J. OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera. Brain Sciences. 2022; 12(1):100. https://doi.org/10.3390/brainsci12010100
Chicago/Turabian StyleTowner, Rheal A., James Hocker, Nataliya Smith, Debra Saunders, James Battiste, and Jay Hanas. 2022. "OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera" Brain Sciences 12, no. 1: 100. https://doi.org/10.3390/brainsci12010100
APA StyleTowner, R. A., Hocker, J., Smith, N., Saunders, D., Battiste, J., & Hanas, J. (2022). OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera. Brain Sciences, 12(1), 100. https://doi.org/10.3390/brainsci12010100