Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects of Study
2.2. Nun Study Design
2.3. Isolation of Mononuclear Cells
2.4. Reverse Transcription
2.5. Quantification by MassARRAY Quantitative Gene Expression (QGE) Analysis
2.6. Determination of Plasma Biomarkers
2.7. Determination of Urinary Biomarkers
2.8. Storage, Preparation, and 1H NMR Spectroscopic Analysis of Blood Plasma for Metabolomics
2.9. Multivariate Analysis of Metabolomic NMR Spectra
2.10. Life Span in Drosophila melanogaster
2.11. Food Intake Determination in Drosophila melanogaster
2.12. RNA Extraction and the Real-Time RT Polymerase Chain Reaction (RT-PCR) in Drosophila melanogaster
2.13. Statistical Analysis
3. Results
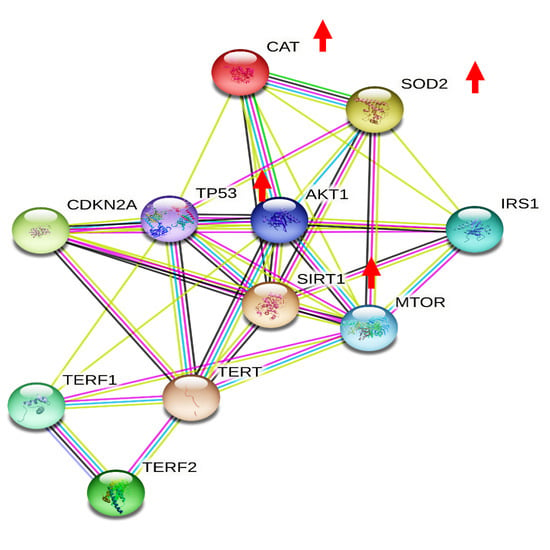
3.1. Effects of Moderate Red Wine Consumption on mRNA Levels of Longevity-Related Genes in Human Blood Mononuclear Cells
3.2. Effect of Red Wine Consumption on Markers of Oxidative Stress and Levels of Antioxidant Vitamins
3.3. Metabolomic Profile after Red Wine Consumption Suggest Better Cardiometabolic Health
3.4. Lack of Adverse Effects of Moderate Red Wine Consumption on General Biochemical and Clinical Parameters
3.5. Wine Consumption Increases Life Span and Upregulates Longevity-Related Gene Expression in Controlled Populations of Drosophila melanogaster
4. Discussion
4.1. The J-Shaped Curve of the Effects of Wine Consumption on Health
4.2. The Effect of Red Wine in Controlled Populations
4.3. Should We Recommend the Consumption of Small Amounts of Wine to the General Population?
4.4. Limitations of the Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jouanna, J. Greek Medicine from Hippocrates to Galen: Selected Papers; Brill: Leiden, The Netherlands, 2012; ISBN 978-90-04-23254-9. [Google Scholar]
- Patay, R.; Danon, G. [On some biological changes produced, in the swine, by the daily consumption of red wine]. Bull. Acad. Natl. Med. 1962, 146, 62–65. [Google Scholar] [PubMed]
- Renaud, S.; de Lorgeril, M. Wine, Alcohol, Platelets, and the French Paradox for Coronary Heart Disease. Lancet 1992, 339, 1523–1526. [Google Scholar] [CrossRef]
- Richard, J.L. [Coronary risk factors. The French paradox]. Arch. Mal. Coeur. Vaiss. 1987, 80, 17–21. [Google Scholar]
- Lavy, A.; Fuhrman, B.; Markel, A.; Dankner, G.; Ben-Amotz, A.; Presser, D.; Aviram, M. Effect of Dietary Supplementation of Red or White Wine on Human Blood Chemistry, Hematology and Coagulation: Favorable Effect of Red Wine on Plasma High-Density Lipoprotein. Ann. Nutr. Metab. 1994, 38, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, J.; Shearer, G.C.; Lichtenstein, A.H.; Zheng, X.; Wu, Y.; Jin, C.; Wu, S.; Gao, X. Longitudinal Study of Alcohol Consumption and HDL Concentrations: A Community-Based Study. Am. J. Clin. Nutr. 2017, 105, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayek, T.; Fuhrman, B.; Vaya, J.; Rosenblat, M.; Belinky, P.; Coleman, R.; Elis, A.; Aviram, M. Reduced Progression of Atherosclerosis in Apolipoprotein E-Deficient Mice Following Consumption of Red Wine, or Its Polyphenols Quercetin or Catechin, Is Associated with Reduced Susceptibility of LDL to Oxidation and Aggregation. Arterioscler. Thromb. Vasc. Biol. 1997, 17, 2744–2752. [Google Scholar] [CrossRef]
- Aviram, M.; Hayek, T.; Fuhrman, B. Red Wine Consumption Inhibits LDL Oxidation and Aggregation in Humans and in Atherosclerotic Mice. Biofactors 1997, 6, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Pinotti, F.; Wikramaratna, P.S.; Obolski, U.; Paton, R.S.; Damineli, D.S.C.; Alcantara, L.C.J.; Giovanetti, M.; Gupta, S.; Lourenço, J. Potential Impact of Individual Exposure Histories to Endemic Human Coronaviruses on Age-Dependent Severity of COVID-19. BMC Med. 2021, 19, 19. [Google Scholar] [CrossRef]
- Chen, W.-M.; Shaw, L.-H.; Chang, P.-J.; Tung, S.-Y.; Chang, T.-S.; Shen, C.-H.; Hsieh, Y.-Y.; Wei, K.-L. Hepatoprotective Effect of Resveratrol against Ethanol-Induced Oxidative Stress through Induction of Superoxide Dismutase in Vivo and in Vitro. Exp. Ther. Med. 2016, 11, 1231–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorelik, S.; Ligumsky, M.; Kohen, R.; Kanner, J. A Novel Function of Red Wine Polyphenols in Humans: Prevention of Absorption of Cytotoxic Lipid Peroxidation Products. FASEB J. 2008, 22, 41–46. [Google Scholar] [CrossRef]
- Magyar, K.; Halmosi, R.; Palfi, A.; Feher, G.; Czopf, L.; Fulop, A.; Battyany, I.; Sumegi, B.; Toth, K.; Szabados, E. Cardioprotection by Resveratrol: A Human Clinical Trial in Patients with Stable Coronary Artery Disease. Clin. Hemorheol. Microcirc. 2012, 50, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Semba, R.D.; Ferrucci, L.; Bartali, B.; Urpí-Sarda, M.; Zamora-Ros, R.; Sun, K.; Cherubini, A.; Bandinelli, S.; Andres-Lacueva, C. Resveratrol Levels and All-Cause Mortality in Older Community-Dwelling Adults. JAMA Intern. Med. 2014, 174, 1077–1084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tresserra-Rimbau, A.; Medina-Remón, A.; Lamuela-Raventós, R.M.; Bulló, M.; Salas-Salvadó, J.; Corella, D.; Fitó, M.; Gea, A.; Gómez-Gracia, E.; Lapetra, J.; et al. Moderate Red Wine Consumption Is Associated with a Lower Prevalence of the Metabolic Syndrome in the PREDIMED Population. Br. J. Nutr. 2015, 113 (Suppl. 2), S121–S130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, S.R.; Rendeiro, C.; McGettrick, H.M.; Philp, A.; Lucas, S.J.E. Fine Wine or Sour Grapes? A Systematic Review and Meta-Analysis of the Impact of Red Wine Polyphenols on Vascular Health. Eur. J. Nutr. 2020. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Podda, M.; Rallis, M.; Traber, M.G.; Packer, L.; Maibach, H.I. Kinetic Study of Cutaneous and Subcutaneous Distribution Following Topical Application of [7,8-14C]Rac-Alpha-Lipoic Acid onto Hairless Mice. Biochem. Pharmacol. 1996, 52, 627–633. [Google Scholar] [CrossRef]
- Taylor, A.W.; Bruno, R.S.; Traber, M.G. Women and Smokers Have Elevated Urinary F(2)-Isoprostane Metabolites: A Novel Extraction and LC-MS Methodology. Lipids 2008, 43, 925–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, S.W.; Traber, M.G. Measurement of the Vitamin E Metabolites, Carboxyethyl Hydroxychromans (CEHCs), in Biological Samples. Curr. Protoc. Toxicol. 2006, 29, 7.8.1–7.8.12. [Google Scholar] [CrossRef]
- Hervey, G.R. Determination of Creatinine by the Jaffé Reaction. Nature 1953, 171, 1125. [Google Scholar] [CrossRef] [PubMed]
- Orr, W.C.; Mockett, R.J.; Benes, J.J.; Sohal, R.S. Effects of Overexpression of Copper-Zinc and Manganese Superoxide Dismutases, Catalase, and Thioredoxin Reductase Genes on Longevity in Drosophila melanogaster. J. Biol. Chem. 2003, 278, 26418–26422. [Google Scholar] [CrossRef] [Green Version]
- Borrás, C.; Sastre, J.; García-Sala, D.; Lloret, A.; Pallardó, F.V.; Viña, J. Mitochondria from Females Exhibit Higher Antioxidant Gene Expression and Lower Oxidative Damage than Males. Free Radic. Biol. Med. 2003, 34, 546–552. [Google Scholar] [CrossRef]
- Calleja, M.; Peña, P.; Ugalde, C.; Ferreiro, C.; Marco, R.; Garesse, R. Mitochondrial DNA Remains Intact during Drosophila Aging, but the Levels of Mitochondrial Transcripts Are Significantly Reduced. J. Biol. Chem. 1993, 268, 18891–18897. [Google Scholar] [CrossRef]
- Crawford, D.R.; Wang, Y.; Schools, G.P.; Kochheiser, J.; Davies, K.J. Down-Regulation of Mammalian Mitochondrial RNAs during Oxidative Stress. Free Radic. Biol. Med. 1997, 22, 551–559. [Google Scholar] [CrossRef]
- Kloner, R.A.; Rezkalla, S.H. To Drink or Not to Drink? That Is the Question. Circulation 2007, 116, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.M.; Kaptoge, S.; Butterworth, A.S.; Willeit, P.; Warnakula, S.; Bolton, T.; Paige, E.; Paul, D.S.; Sweeting, M.; Burgess, S.; et al. Risk Thresholds for Alcohol Consumption: Combined Analysis of Individual-Participant Data for 599 912 Current Drinkers in 83 Prospective Studies. Lancet 2018, 391, 1513–1523. [Google Scholar] [CrossRef] [Green Version]
- Calabrese, V.; Cornelius, C.; Dinkova-Kostova, A.T.; Calabrese, E.J.; Mattson, M.P. Cellular Stress Responses, the Hormesis Paradigm, and Vitagenes: Novel Targets for Therapeutic Intervention in Neurodegenerative Disorders. Antioxid. Redox Signal. 2010, 13, 1763–1811. [Google Scholar] [CrossRef] [PubMed]
- Siracusa, R.; Scuto, M.; Fusco, R.; Trovato, A.; Ontario, M.L.; Crea, R.; Di Paola, R.; Cuzzocrea, S.; Calabrese, V. Anti-Inflammatory and Anti-Oxidant Activity of Hidrox® in Rotenone-Induced Parkinson’s Disease in Mice. Antioxidants 2020, 9, 824. [Google Scholar] [CrossRef]
- Kołota, A.; Głąbska, D.; Oczkowski, M.; Gromadzka-Ostrowska, J. Analysis of Association between Intake of Red Wine Polyphenols and Oxidative Stress Parameters in the Liver of Growing Male Rats. Appl. Sci. 2020, 10, 6389. [Google Scholar] [CrossRef]
- Di Renzo, L.; Carraro, A.; Valente, R.; Iacopino, L.; Colica, C.; De Lorenzo, A. Intake of Red Wine in Different Meals Modulates Oxidized LDL Level, Oxidative and Inflammatory Gene Expression in Healthy People: A Randomized Crossover Trial. Available online: https://www.hindawi.com/journals/omcl/2014/681318/ (accessed on 31 January 2021).
- Ahola-Olli, A.V.; Mustelin, L.; Kalimeri, M.; Kettunen, J.; Jokelainen, J.; Auvinen, J.; Puukka, K.; Havulinna, A.S.; Lehtimäki, T.; Kähönen, M.; et al. Circulating Metabolites and the Risk of Type 2 Diabetes: A Prospective Study of 11,896 Young Adults from Four Finnish Cohorts. Diabetologia 2019, 62, 2298–2309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.-Y.; Monleon, D.; Verhamme, P.; Staessen, J.A. Branched-Chain Amino Acids as Critical Switches in Health and Disease. Hypertension 2018, 72, 1012–1022. [Google Scholar] [CrossRef]
- Gonzalez-Freire, M.; Adelnia, F.; Moaddel, R.; Ferrucci, L. Searching for a Mitochondrial Root to the Decline in Muscle Function with Ageing. J. Cachexia Sarcopenia Muscle 2018, 9, 435–440. [Google Scholar] [CrossRef] [Green Version]
- Puchalska, P.; Crawford, P.A. Multi-Dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef] [Green Version]
- Zeisel, S.H.; da Costa, K.-A. Choline: An Essential Nutrient for Public Health. Nutr. Rev. 2009, 67, 615–623. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.-J.; Arneja, A.S.; Tappia, P.S.; Dhalla, N.S. The Potential Health Benefits of Taurine in Cardiovascular Disease. Exp. Clin. Cardiol. 2008, 13, 57–65. [Google Scholar]
- Li, Y.; Cao, Z.; Zhu, H. Upregulation of Endogenous Antioxidants and Phase 2 Enzymes by the Red Wine Polyphenol, Resveratrol in Cultured Aortic Smooth Muscle Cells Leads to Cytoprotection against Oxidative and Electrophilic Stress. Pharmacol. Res. 2006, 53, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Puiggros, F.; Llópiz, N.; Ardévol, A.; Bladé, C.; Arola, L.; Salvadó, M.J. Grape Seed Procyanidins Prevent Oxidative Injury by Modulating the Expression of Antioxidant Enzyme Systems. J. Agric. Food Chem. 2005, 53, 6080–6086. [Google Scholar] [CrossRef]
- Gambini, J.; Gomez-Cabrera, M.C.; Borras, C.; Valles, S.L.; Lopez-Grueso, R.; Martinez-Bello, V.E.; Herranz, D.; Pallardo, F.V.; Tresguerres, J.A.F.; Serrano, M.; et al. Free [NADH]/[NAD(+)] Regulates Sirtuin Expression. Arch. Biochem. Biophys. 2011, 512, 24–29. [Google Scholar] [CrossRef]
- Borrás, C.; Gambini, J.; Gómez-Cabrera, M.C.; Sastre, J.; Pallardó, F.V.; Mann, G.E.; Viña, J. Genistein, a Soy Isoflavone, up-Regulates Expression of Antioxidant Genes: Involvement of Estrogen Receptors, ERK1/2, and NFkappaB. FASEB J. 2006, 20, 2136–2138. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhao, Y. Absorption and Metabolism of Red Wine Polyphenols and Their Potential Health Benefits in Cardiovascular Function. Am. J. Clin. Nutr. 2012, 95, 1496–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pan, A.; Wang, D.D.; Liu, X.; Dhana, K.; Franco, O.H.; Kaptoge, S.; Di Angelantonio, E.; Stampfer, M.; Willett, W.C.; et al. Impact of Healthy Lifestyle Factors on Life Expectancies in the US Population. Circulation 2018, 138, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Loef, M.; Walach, H. The Combined Effects of Healthy Lifestyle Behaviors on All Cause Mortality: A Systematic Review and Meta-Analysis. Prev. Med. 2012, 55, 163–170. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gambini, J.; Gimeno-Mallench, L.; Olaso-Gonzalez, G.; Mastaloudis, A.; Traber, M.G.; Monleón, D.; Borrás, C.; Viña, J. Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster. Antioxidants 2021, 10, 301. https://doi.org/10.3390/antiox10020301
Gambini J, Gimeno-Mallench L, Olaso-Gonzalez G, Mastaloudis A, Traber MG, Monleón D, Borrás C, Viña J. Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster. Antioxidants. 2021; 10(2):301. https://doi.org/10.3390/antiox10020301
Chicago/Turabian StyleGambini, Juan, Lucia Gimeno-Mallench, Gloria Olaso-Gonzalez, Angela Mastaloudis, Maret G. Traber, Daniel Monleón, Consuelo Borrás, and Jose Viña. 2021. "Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster" Antioxidants 10, no. 2: 301. https://doi.org/10.3390/antiox10020301
APA StyleGambini, J., Gimeno-Mallench, L., Olaso-Gonzalez, G., Mastaloudis, A., Traber, M. G., Monleón, D., Borrás, C., & Viña, J. (2021). Moderate Red Wine Consumption Increases the Expression of Longevity-Associated Genes in Controlled Human Populations and Extends Lifespan in Drosophila melanogaster. Antioxidants, 10(2), 301. https://doi.org/10.3390/antiox10020301