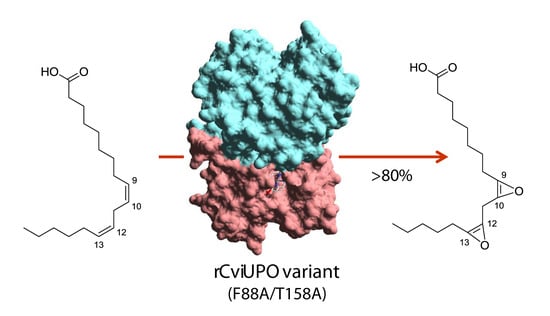
Engineering Collariella virescens Peroxygenase for Epoxides Production from Vegetable Oil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Native Enzyme and Site-Directed Variants
2.2. Enzyme Kinetics
2.3. Epoxidation of Individual Fatty Acids and Oil Hydrolyzate
2.4. GC-MS Analyses
2.5. Chiral Analyses
3. Results and Discussion
3.1. Design and Catalytic Characterization of rCviUPO Variants
3.2. Fatty-Acid Oxygenation Patterns by rCviUPO and Heme-Channel Variants
3.3. Optimization of Hydrolyzed Sunflower Oil Epoxidation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, Y.; Taddeo, F.; Aguilera, A.F.; Cai, X.; Russo, V.; Tolvanen, P.; Leveneur, S. The Lord of the chemical rings: Catalytic synthesis of important industrial epoxide compounds. Catalysts 2021, 11, 7. [Google Scholar] [CrossRef]
- Biermann, U.; Friedt, W.; Lang, S.; Lühs, W.; Machmüller, G.; Metzger, U.O.; Klaas, M.R.; Schäfer, H.J.; Schneider, M.P. New syntheses with oils and fats as renewable raw materials for the chemical industry. In Biorefineries-Industrial Processes and Products; Kamm, B., Gruber, P.R., Kamm, M., Eds.; Wiley-VCH Verlag GmbH: Weinheim, Germany, 2006; pp. 253–289. [Google Scholar]
- Corma, A.; Iborra, S.; Velty, A. Chemical routes for the transformation of biomass into chemicals. Chem. Rev. 2007, 107, 2411–2502. [Google Scholar] [CrossRef] [PubMed]
- Biermann, U.; Bornscheuer, U.; Meier, M.A.R.; Metzger, J.O.; Schäfer, H.J. Oils and Fats as Renewable Raw Materials in Chemistry. Angew. Chem. Int. Ed. 2011, 50, 3854–3871. [Google Scholar] [CrossRef] [PubMed]
- Prileschajew, N. Oxydation ungesättigter Verbindungen mittels organischer Superoxyde. Ber. Dtsch. Chem. Ges. 1909, 42, 4811–4815. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.L.; Wärnå, J.; Salmi, T.; Burel, F.; Taouk, B.; Leveneur, S. Kinetic modeling strategy for an exothermic multiphase reactor system: Application to vegetable oils epoxidation using Prileschajew method. AIChE J. 2016, 62, 726–741. [Google Scholar] [CrossRef]
- Santacesaria, E.; Tesser, R.; Di Serio, M.; Turco, R.; Russo, V.; Verde, D. A biphasic model describing soybean oil epoxidation with H2O2 in a fed-batch reactor. Chem. Eng. J. 2011, 173, 198–209. [Google Scholar] [CrossRef]
- Sinadinovic-Fiser, S.; Jankovic, M.; Borota, O. Epoxidation of castor oil with peracetic acid formed in situ in the presence of an ion exchange resin. Chem. Eng. Process. 2012, 62, 106–113. [Google Scholar] [CrossRef]
- Björkling, F.; Frykman, H.; Godtfredsen, S.E.; Kirk, O. Lipase catalyzed synthesis of peroxycarboxylic acids and lipase mediated oxidations. Tetrahedron 1992, 48, 4587–4592. [Google Scholar] [CrossRef]
- Aouf, C.; Durand, E.; Lecomte, J.; Figueroa-Espinoza, M.C.; Dubreucq, E.; Fulcrand, H.; Villeneuve, P. The use of lipases as biocatalysts for the epoxidation of fatty acids and phenolic compounds. Green Chem. 2014, 16, 1740–1754. [Google Scholar] [CrossRef] [Green Version]
- Schneider, R.d.C.; Lara, L.R.S.; Bitencourt, T.B.; Nascimento, M.d.G.; Nunes, M.R. Chemo-enzymatic epoxidation of sunflower oil methyl esters. J. Braz. Chem. Soc. 2009, 20, 1473–1477. [Google Scholar] [CrossRef]
- Vlcek, T.; Petrovic, Z.S. Optimization of the chemoenzymatic epoxidation of soybean oil. J. Am. Oil Chem. Soc. 2006, 83, 247–252. [Google Scholar] [CrossRef]
- Rüsch gen. Klaas, M.; Warwel, S. Complete and partial epoxidation of plant oils by lipase-catalyzed perhydrolysis. Ind. Crops Prod. 1999, 9, 125–132. [Google Scholar] [CrossRef]
- Rüsch gen. Klaas, M.; Warwel, S. Lipase-catalyzed preparation of peroxy acids and their use for epoxidation. J. Mol. Catal. A Chem. 1997, 117, 311–319. [Google Scholar] [CrossRef]
- Piazza, G.J.; Nuñez, A.; Foglia, T.A. Epoxidation of fatty acids, fatty methyl esters, and alkenes by immobilized oat seed peroxygenase. J. Mol. Catal. B-Enzym. 2003, 21, 143–151. [Google Scholar] [CrossRef]
- Oliw, E.H. Oxygenation of polyunsaturated fatty acids by cytochrome P450 monooxygenases. Progr. Lipid Res. 1994, 33, 329–354. [Google Scholar] [CrossRef]
- Aranda, C.; Olmedo, A.; Kiebist, J.; Scheibner, K.; del Río, J.C.; Martínez, A.T.; Gutiérrez, A. Selective epoxidation of fatty acids and fatty acid methyl esters by fungal peroxygenases. ChemCatChem 2018, 10, 3964–3968. [Google Scholar] [CrossRef] [Green Version]
- Hofrichter, M.; Kellner, H.; Pecyna, M.J.; Ullrich, R. Fungal unspecific peroxygenases: Heme-thiolate proteins that combine peroxidase and cytochrome P450 properties. Adv. Exp. Med. Biol. 2015, 851, 341–368. [Google Scholar]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Liers, C.; Scheibner, K.; Wambui, V.; Ullrich, R. Fungal peroxygenases: A phylogenetically old superfamily of heme enzymes with promiscuity for oxygen transfer reactions. In Grand Challenges in Fungal Biotechnology; Nevalainen, H., Ed.; Springer: Cham, Switzerland, 2020; pp. 369–403. [Google Scholar]
- Ullrich, R.; Hofrichter, M. Enzymatic hydroxylation of aromatic compounds. Cell. Mol. Life Sci. 2007, 64, 271–293. [Google Scholar] [CrossRef]
- Gutiérrez, A.; Babot, E.D.; Ullrich, R.; Hofrichter, M.; Martínez, A.T.; del Río, J.C. Regioselective oxygenation of fatty acids, fatty alcohols and other aliphatic compounds by a basidiomycete heme-thiolate peroxidase. Arch. Biochem. Biophys. 2011, 514, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Peter, S.; Kinne, M.; Wang, X.; Ulrich, R.; Kayser, G.; Groves, J.T.; Hofrichter, M. Selective hydroxylation of alkanes by an extracellular fungal peroxygenase. FEBS J. 2011, 278, 3667–3675. [Google Scholar] [CrossRef]
- Aranda, C.; Carro, J.; González-Benjumea, A.; Babot, E.D.; Olmedo, A.; Linde, D.; Martínez, A.T.; Gutiérrez, A. Advances in enzymatic oxyfunctionalization of aliphatic compounds. Biotechnol. Adv. 2021, 51, 107703. [Google Scholar] [PubMed]
- Municoy, M.; González-Benjumea, A.; Carro, J.; Aranda, C.; Linde, D.; Renau-Mínguez, C.; Ullrich, R.; Hofrichter, M.; Guallar, V.; Gutiérrez, A.; et al. Fatty-acid oxygenation by fungal peroxygenases: From computational simulations to preparative regio- and stereo-selective epoxidation. ACS Catal. 2020, 10, 13584–13595. [Google Scholar] [CrossRef]
- González-Benjumea, A.; Carro, J.; Renau, C.; Linde, D.; Fernández-Fueyo, E.; Gutiérrez, A.; Martínez, A.T. Fatty acid epoxidation by Collariella virescens peroxygenase and heme-channel variants. Catal. Sci. Technol. 2020, 10, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Carro, J.; González-Benjumea, A.; Fernández-Fueyo, E.; Aranda, C.; Guallar, V.; Gutiérrez, A.; Martínez, A.T. Modulating fatty acid epoxidation vs hydroxylation in a fungal peroxygenase. ACS Catal. 2019, 9, 6234–6242. [Google Scholar] [CrossRef] [Green Version]
- Babot, E.D.; del Río, J.C.; Kalum, L.; Martínez, A.T.; Gutiérrez, A. Oxyfunctionalization of aliphatic compounds by a recombinant peroxygenase from Coprinopsis cinerea. Biotechnol. Bioeng. 2013, 110, 2332. [Google Scholar]
- Linde, D.; Olmedo, A.; González-Benjumea, A.; Renau, C.; Estévez, M.; Carro, J.; Fernández-Fueyo, E.; Gutiérrez, A.; Martínez, A.T. Two new unspecific peroxygenases from heterologous expression of fungal genes in Escherichia coli. Appl. Environ. Microbiol. 2020, 86, e02899-19. [Google Scholar] [CrossRef] [Green Version]
- González-Benjumea, A.; Linde, D.; Carro, J.; Ullrich, R.; Hofrichter, M.; Martínez, A.T.; Gutiérrez, A. Regioselective and stereoselective epoxidation of n-3 and n-6 fatty acids by fungal peroxygenases. Antioxidants 2021, 10, 1888. [Google Scholar]
- Fernández-Fueyo, E.; Aranda, C.; Gutiérrez, A.; Martínez, A.T. Method of Heterologous Expression of Active Fungal Unspecific Peroxygenase in Bacterial Host Cells for Fatty-Acid Epoxidation and Other Oxygenation Reactions. European Patent EP18382514.0, 10 July 2018. [Google Scholar]
- Lund, H.; Kalum, L.; Hofrichter, M.; Peter, S. Epoxidation Using Peroxygenase. U.S. Patent 9908860 B2, 6 March 2018. [Google Scholar]
- Puigbò, P.; Guzmán, E.; Romeu, A.; Garcia-Vallvé, S. OPTIMIZER: A web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007, 35, W126–W131. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar]
- Ullrich, R.; Nuske, J.; Scheibner, K.; Spantzel, J.; Hofrichter, M. Novel haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 2004, 70, 4575–4581. [Google Scholar]
- Piontek, K.; Strittmatter, E.; Ullrich, R.; Gröbe, G.; Pecyna, M.J.; Kluge, M.; Scheibner, K.; Hofrichter, M.; Plattner, D.A. Structural basis of substrate conversion in a new aromatic peroxygenase: Cytochrome P450 functionality with benefits. J. Biol. Chem. 2013, 288, 34767–34776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- González-Benjumea, A.; Marques, G.; Herold-Majumdar, O.M.; Kiebist, J.; Scheibner, K.; del Río, J.C.; Martínez, A.T.; Gutiérrez, A. High epoxidation yields of vegetable oil hydrolyzates and methyl esters by selected fungal peroxygenases. Front. Bioeng. Biotechnol. 2021, 8, 605854. [Google Scholar] [CrossRef] [PubMed]
- Pinot, F.; Benveniste, I.; Salaün, J.P.; Loreau, O.; Noël, J.P.; Schreiber, L.; Durst, F. Production in vitro by the cytochrome P450 CYP94A1 of major C18 cutin monomers and potential messengers in plant-pathogen interactions: Enantioselectivity studies. Biochem. J. 1999, 342, 27–32. [Google Scholar] [CrossRef]
- Bormann, S.; Baraibar, A.G.; Ni, Y.; Holtmann, D.; Hollmann, F. Specific oxyfunctionalisations catalysed by peroxygenases: Opportunities, challenges and solutions. Catal. Sci. Technol. 2015, 5, 2038–2052. [Google Scholar] [CrossRef] [Green Version]
- Faiza, M.; Huang, S.F.; Lan, D.M.; Wang, Y.H. New insights on unspecific peroxygenases: Superfamily reclassification and evolution. BMC Evol. Biol. 2019, 19, 76. [Google Scholar] [CrossRef] [Green Version]
- Grogan, G. Hemoprotein catalyzed oxygenations: P450s, UPOs, and progress toward scalable reactions. JACS Au 2021, 1, 1312–1329. [Google Scholar] [CrossRef]
- Hobisch, M.; Holtmann, D.; Gomez de Santos, P.; Alcalde, M.; Hollmann, F.; Kara, S. Recent developments in the use of peroxygenases- Exploring their high potential in selective oxyfunctionalisations. Biotechnol. Adv. 2021, 51, 107615. [Google Scholar] [CrossRef]
- Münch, J.; Püllmann, P.; Zhang, W.; Weissenborn, M.J. Enzymatic hydroxylations of sp3-carbons. ACS Catal. 2021, 11, 9168–9203. [Google Scholar] [CrossRef]
- Beltrán-Nogal, A.; Sánchez-Moreno, I.; Méndez-Sánchez, D.; Gómez de Santos, P.; Hollmann, F.; Alcalde, M. Surfing the wave of oxyfunctionalization chemistry by engineering fungal unspecific peroxygenases. Curr. Opin. Struct. Biol. 2022, 73, 102342. [Google Scholar] [CrossRef]
- Hofrichter, M.; Kellner, H.; Herzog, R.; Karich, A.; Kiebist, J.; Scheibner, K.; Ullrich, R. Peroxide-mediated oxygenation of organic compounds by fungal peroxygenases. Antioxidants 2022, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lan, D.; Durrani, R.; Hollmann, F. Peroxygenases en route to becoming dream catalysts. What are the opportunities and challenges? Curr. Opin. Chem. Biol. 2017, 37, 1–9. [Google Scholar] [CrossRef] [PubMed]




| Km (µM) | kcat/Km (s−1·mM−1) | ki (µM) | |
|---|---|---|---|
| Native | 1600 ± 468 | 374 ± 129 | 5580 ± 1800 |
| F88A | 16,300 ± 2900 | 29 ± 6 | - |
| T158A | 23,800 ± 13,700 | 127 ± 99 | 1790 ± 1000 |
| F88A/T158A | 9370 ± 880 | 55 ± 6 | - |
| 6Ala | 14,800 ± 1600 | 0.7 ± 0.1 | - |
| kcat (s−1) | Km (µM) | kcat/Km (s−1·mM−1) | ki (µM) | |
|---|---|---|---|---|
| Native | 12.6 ± 0.3 | 58.8 ± 6.0 | 214 ± 53 | - |
| F88A | 10.5 ± 1.7 | 278.0 ± 49.0 | 38 ± 9 | 1120 ± 400 |
| T158A | 21.0 ± 1.1 | 89.0 ± 16.4 | 184 ± 35 | - |
| F88A/T158A | 16.5 ± 0.5 | 51.6 ± 5.4 | 319 ± 35 | 2900 ± 610 |
| 6Ala | 16.6 ± 3.6 | 399.0 ± 94.6 | 42 ± 13 | 1360 ± 710 |
| Conversion | Products | (%) | Epoxidation | |||||
|---|---|---|---|---|---|---|---|---|
| (%) | 15-Epoxy | 12-Epoxy | 9-Epoxy | Diepoxy | OH/Keto | OH-Epoxy | Yield (%) | |
| C18:1 | ||||||||
| Native | 96 | - | - | 71 | - | 28 | 1 | 71 |
| F88A | 97 | - | - | 69 | - | 6 | 25 | 91 |
| T158A | 98 | - | - | 87 | - | 5 | 8 | 93 |
| F88A/T158A | 95 | - | - | 63 | - | 13 | 24 | 82 |
| 6Ala | 96 | - | - | 96 | - | 4 | - | 92 |
| C18:2 | ||||||||
| Native | 97 | - | 56 | 10 | - | 8 | 26 | 45 |
| F88A | 98 | - | 15 | 2 | 46 | 12 | 25 | 66 |
| T158A | 88 | - | 23 | 17 | 29 | 27 | 4 | 43 |
| F88A/T158A | 99 | - | 4 | - | 81 | - | 15 | 90 |
| 6Ala | 99 | - | - | 25 | 64 | - | 11 | 81 |
| C18:3 | ||||||||
| Native | 96 | 77 | 6 | 2 | - | - | 15 | 32 |
| F88A | 98 | 16 | 6 | 4 | 53 | 8 | 13 | 47 |
| T158A | 98 | 26 | 30 | 17 | 3 | 20 | 3 | 27 |
| F88A/T158A | 99 | 2 | 3 | - | 82 | - | 13 | 60 |
| 6Ala | 99 | 17 | 35 | 16 | 10 | 16 | 6 | 47 |
| Entry | Preparation | Substrate, mM | Enzyme, µM (S/E Ratio) | Acetone, % | H2O2, mM (Equiv) | Time, min | Epoxidation Yield, % |
|---|---|---|---|---|---|---|---|
| 1 | native | 0.1 | 0.25 (400) | 20 | 1 (6.8) | 30 | 54 |
| 2 | native | 0.1 | 0.5 (200) | 20 | 1 (6.8) | 30 | 52 |
| 3 | native | 0.1 | 1 (100) | 20 | 1 (6.8) | 30 | 53 |
| 4 | F88A/T158A | 0.1 | 0.25 (400) | 20 | 1 (6.8) | 30 | 59 |
| 5 | F88A/T158A | 0.1 | 0.5 (200) | 20 | 1 (6.8) | 30 | 72 |
| 6 | F88A/T158A | 0.1 | 1 (100) | 20 | 1 (6.8) | 30 | 75 |
| 7 | native | 5 | 12.5 (400) | 20 | 50 (6.8) | 30 | 8 |
| 8 | native | 5 | 12.5 (400) | 20 | 50 (6.8) | 60 | 28 |
| 9 | F88A/T158A | 5 | 25 (200) | 20 | 50 (6.8) | 30 | 35 |
| 10 | F88A/T158A | 5 | 25 (200) | 20 | 50 (6.8) | 60 | 84 |
| 11 | native | 10 | 25 (400) | 20 | 100 (6.8) | 60 | 37 |
| 12 | native | 10 | 25 (400) | 30 | 100 (6.8) | 60 | 56 |
| 13 | F88A/T158A | 10 | 50 (200) | 20 | 100 (6.8) | 60 | 40 |
| 14 | F88A/T158A | 10 | 50 (200) | 30 | 100 (6.8) | 60 | 79 |
| 15 | F88A/T158A | 10 | 50 (200) | 30 | 50 (3.4) | 60 | 78 |
| 16 | F88A/T158A | 10 | 50 (200) | 30 | 25 (1.7) | 60 | 85 |
| 17 | F88A/T158A | 10 | 50 (200) | 30 | 15 (1.0) | 60 | 44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linde, D.; González-Benjumea, A.; Aranda, C.; Carro, J.; Gutiérrez, A.; Martínez, A.T. Engineering Collariella virescens Peroxygenase for Epoxides Production from Vegetable Oil. Antioxidants 2022, 11, 915. https://doi.org/10.3390/antiox11050915
Linde D, González-Benjumea A, Aranda C, Carro J, Gutiérrez A, Martínez AT. Engineering Collariella virescens Peroxygenase for Epoxides Production from Vegetable Oil. Antioxidants. 2022; 11(5):915. https://doi.org/10.3390/antiox11050915
Chicago/Turabian StyleLinde, Dolores, Alejandro González-Benjumea, Carmen Aranda, Juan Carro, Ana Gutiérrez, and Angel T. Martínez. 2022. "Engineering Collariella virescens Peroxygenase for Epoxides Production from Vegetable Oil" Antioxidants 11, no. 5: 915. https://doi.org/10.3390/antiox11050915
APA StyleLinde, D., González-Benjumea, A., Aranda, C., Carro, J., Gutiérrez, A., & Martínez, A. T. (2022). Engineering Collariella virescens Peroxygenase for Epoxides Production from Vegetable Oil. Antioxidants, 11(5), 915. https://doi.org/10.3390/antiox11050915