Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae
Abstract
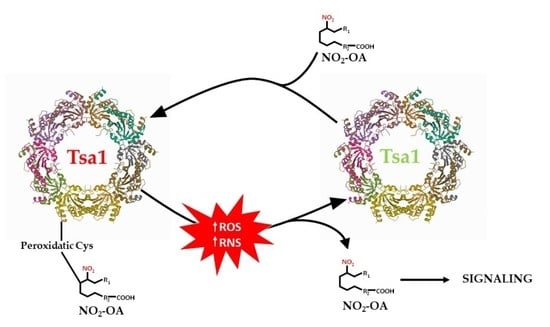
:1. Introduction
2. Materials and Methods
2.1. Organisms, Liquid Medium and Growth Conditions
2.2. Heat Stress Conditions
2.3. Obtaining the Cell-Free Extract
2.4. Quantification of Hydrogen Peroxide (H2O2)
2.5. Enzymatic Peroxiredoxin Activity Assay
2.6. Immunodetection of Tsa1
2.7. Synthesis and Characterization of the NO2-OA Standard and the 13C18-NO2-OA Internal Standard by NMR Spectroscopy
2.8. Lipid Extraction and Acid Hydrolysis
2.9. Detection and Identification of Endogenous NO2-OA in Saccharomyces Cerevisiae
2.10. Expression and Purification of Recombinant Tsa1
2.11. In Vitro Nitroalkylation of Recombinant Tsa1
2.12. Extraction of the In Vivo Nitroalkylated Peptides
2.13. Detection of Protein Nitroalkylation by nano-LC-MS/MS
2.14. MS/MS Data Processing and Identification of Nitroalkylated Proteins
2.15. Relative Quantification of Tsa1 Nitroalkylation by nano-LC-MS/MS
2.16. Docking
3. Results
3.1. Heat Stress Activates Tsa1 Enzymatic Activity
3.2. Detection and Quantification of Endogenous NO2-OA Levels
3.3. Effect of NO2-OA on the Enzymatic Activity of Tsa1
3.4. Characterization of the Nitroalkylation of Recombinant Tsa1 from Yeast
3.5. Relative Quantification of the Recombinant Tsa1 Nitroalkylation Targets
3.6. Analysis of the Target Residues Involved in Tsa1 Nitroalkylation
3.7. Detection of the In Vivo Nitroalkylation of Tsa1 in the Control and Stress Situations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamamoto, N.; Maeda, Y.; Ikeda, A.; Sakurai, H. Regulation of thermotolerance by stress-induced transcription factors in Saccharomyces cerevisiae. Eukaryot Cell 2008, 7, 783–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morano, K.A.; Grant, C.M.; Moye-Rowley, W.S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 2012, 190, 1157–1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, Z.A.; Schröder, E.; Robin Harris, J.; Poole, L.B. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003, 28, 32–40. [Google Scholar] [CrossRef]
- Rhee, S.G. Overview on Peroxiredoxin. Mol. Cells 2016, 39, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Toledano, M.B.; Huang, B. Microbial 2-Cys Peroxiredoxins: Insights into Their Complex Physiological Roles. Mol. Cells 2016, 39, 31–39. [Google Scholar] [CrossRef] [Green Version]
- Pedrajas, J.R.; McDonagh, B.; Hernández-Torres, F.; Miranda-Vizuete, A.; González-Ojeda, R.; Martínez-Galisteo, E.; Padilla, C.A.; Bárcena, J.A. Glutathione Is the Resolving Thiol for Thioredoxin Peroxidase Activity of 1-Cys Peroxiredoxin Without Being Consumed During the Catalytic Cycle. Antioxid. Redox Signal. 2016, 24, 115–128. [Google Scholar] [CrossRef]
- Li, C.C.; Yang, M.J.; Yang, J.; Kang, M.; Li, T.; He, L.H.; Song, Y.J.; Zhu, Y.B.; Zhao, N.L.; Zhao, C.; et al. Structural and biochemical analysis of 1-Cys peroxiredoxin ScPrx1 from Saccharomyces cerevisiae mitochondria. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129706. [Google Scholar] [CrossRef]
- Bhatt, I.; Tripathi, B.N. Plant peroxiredoxins: Catalytic mechanisms, functional significance and future perspectives. Biotechnol. Adv. 2011, 29, 850–859. [Google Scholar] [CrossRef]
- Schopfer, F.J.; Khoo, N.K.H. Nitro-Fatty Acid Logistics: Formation, Biodistribution, Signaling, and Pharmacology. Trends Endocrinol. Metab. 2019, 30, 505–519. [Google Scholar] [CrossRef]
- Aranda-Caño, L.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Chaki, M.; Mata-Pérez, C.; Padilla, M.N.; Valderrama, R.; Barroso, J.B. Post-Translational Modification of Proteins Mediated by Nitro-Fatty Acids in Plants: Nitroalkylation. Plants 2019, 8, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeman, B.A.; Baker, P.R.; Schopfer, F.J.; Woodcock, S.R.; Napolitano, A.; d’Ischia, M. Nitro-fatty acid formation and signaling. J. Biol. Chem. 2008, 283, 15515–15519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grippo, V.; Mojovic, M.; Pavicevic, A.; Kabelac, M.; Hubatka, F.; Turanek, J.; Zatloukalova, M.; Freeman, B.A.; Vacek, J. Electrophilic characteristics and aqueous behavior of fatty acid nitroalkenes. Redox Biol. 2021, 38, 101756–101768. [Google Scholar] [CrossRef] [PubMed]
- Franz, J.; Bereau, T.; Pannwitt, S.; Anbazhagan, V.; Lehr, A.; Nubbemeyer, U.; Dietz, U.; Bonn, M.; Weidner, T.; Schneider, D. Nitrated Fatty Acids Modulate the Physical Properties of Model Membranes and the Structure of Transmembrane Proteins. Chemistry 2017, 23, 9690–9697. [Google Scholar] [CrossRef] [PubMed]
- Baker, L.M.; Baker, P.R.; Golin-Bisello, F.; Schopfer, F.J.; Fink, M.; Woodcock, S.R.; Branchaud, B.P.; Radi, R.; Freeman, B.A. Nitro-fatty acid reaction with glutathione and cysteine. Kinetic analysis of thiol alkylation by a Michael addition reaction. J. Biol. Chem. 2007, 282, 31085–31093. [Google Scholar] [CrossRef] [Green Version]
- Geisler, A.C.; Rudolph, T.K. Nitroalkylation—A redox sensitive signaling pathway. Biochim. Biophys. Acta 2012, 1820, 777–784. [Google Scholar] [CrossRef]
- Melo, T.; Montero-Bullón, J.F.; Domingues, P.; Domingues, M.R. Discovery of bioactive nitrated lipids and nitro-lipid-protein adducts using mass spectrometry-based approaches. Redox Biol. 2019, 23, 101106–101122. [Google Scholar] [CrossRef]
- Fazzari, M.; Khoo, N.K.; Woodcock, S.R.; Jorkasky, D.K.; Li, L.; Schopfer, F.J.; Freeman, B.A. Nitro-fatty acid pharmacokinetics in the adipose tissue compartment. J. Lipid Res. 2017, 58, 375–385. [Google Scholar] [CrossRef] [Green Version]
- Khoo, N.K.H.; Schopfer, F.J. Nitrated fatty acids: From diet to disease. Curr. Opin. Physiol. 2019, 9, 67–72. [Google Scholar] [CrossRef]
- Rom, O.; Xu, G.; Guo, Y.; Zhu, Y.; Wang, H.; Zhang, J.; Fan, Y.; Liang, W.; Lu, H.; Liu, Y.; et al. Nitro-fatty acids protect against steatosis and fibrosis during development of nonalcoholic fatty liver disease in mice. EBioMedicine 2019, 41, 62–72. [Google Scholar] [CrossRef] [Green Version]
- Turell, L.; Steglich, M.; Alvarez, B. The chemical foundations of nitroalkene fatty acid signaling through addition reactions with thiols. Nitric Oxide 2018, 78, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Vitturi, D.A.; Jorkasky, D.K.; Freeman, B.A. Nitro-fatty acids: New drug candidates for chronic inflammatory and fibrotic diseases. Nitric Oxide 2018, 79, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Piesche, M.; Roos, J.; Kühn, B.; Fettel, J.; Hellmuth, N.; Brat, C.; Maucher, I.V.; Awad, O.; Matrone, C.; Comerma Steffensen, S.G.; et al. The Emerging Therapeutic Potential of Nitro Fatty Acids and Other Michael Acceptor-Containing Drugs for the Treatment of Inflammation and Cancer. Front. Pharmacol. 2020, 11, 1297–1313. [Google Scholar] [CrossRef] [PubMed]
- Panati, K.; Thimmana, L.V.; Narala, V.R. Electrophilic nitrated fatty acids are potential therapeutic candidates for inflammatory and fibrotic lung diseases. Nitric Oxide 2020, 102, 28–38. [Google Scholar] [CrossRef]
- Kansanen, E.; Jyrkkänen, H.K.; Volger, O.L.; Leinonen, H.; Kivelä, A.M.; Häkkinen, S.K.; Woodcock, S.R.; Schopfer, F.J.; Horrevoets, A.J.; Ylä-Herttuala, S.; et al. Nrf2-dependent and -independent responses to nitro-fatty acids in human endothelial cells: Identification of heat shock response as the major pathway activated by nitro-oleic acid. J. Biol. Chem. 2009, 284, 33233–33241. [Google Scholar] [CrossRef] [Green Version]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Luque, F.; Melguizo, M.; Jiménez-Ruiz, J.; Fierro-Risco, J.; Peñas-Sanjuán, A.; Valderrama, R.; et al. Nitro-Fatty Acids in Plant Signaling: Nitro-Linolenic Acid Induces the Molecular Chaperone Network in Arabidopsis. Plant. Physiol. 2016, 170, 686–701. [Google Scholar] [CrossRef]
- Di Fino, L.M.; Cerrudo, I.; Salvatore, S.R.; Schopfer, F.J.; García-Mata, C.; Laxalt, A.M. Exogenous Nitro-Oleic Acid Treatment Inhibits Primary Root Growth by Reducing the Mitosis in the Meristem in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1059–1071. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Padilla, M.N.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Valderrama, R.; Chaki, M.; Aranda-Caño, L.; Moreno-González, D.; Molina-Díaz, A.; Barroso, J.B. Endogenous Biosynthesis of S-Nitrosoglutathione From Nitro-Fatty Acids in Plants. Front. Plant Sci 2020, 11, 962–975. [Google Scholar] [CrossRef]
- Bellincampi, D.; Dipierro, N.; Salvi, G.; Cervone, F.; De Lorenzo, G. Extracellular H2O2 induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol. 2000, 122, 1379–1385. [Google Scholar] [CrossRef] [Green Version]
- Baker, P.R.; Lin, Y.; Schopfer, F.J.; Woodcock, S.R.; Groeger, A.L.; Batthyany, C.; Sweeney, S.; Long, M.H.; Iles, K.E.; Baker, L.M.; et al. Fatty acid transduction of nitric oxide signaling: Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 2005, 280, 42464–42475. [Google Scholar] [CrossRef] [Green Version]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011, 39, W270-7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grosdidier, A.; Zoete, V.; Michielin, O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011, 32, 2149–2159. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2011, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterling, T.; Irwin, J.J. ZINC 15-Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. [Google Scholar] [CrossRef]
- Tairum, C.A.; de Oliveira, M.A.; Horta, B.B.; Zara, F.J.; Netto, L.E.S. Disulfide biochemistry in 2-cys peroxiredoxin: Requirement of Glu50 and Arg146 for the reduction of yeast Tsa1 by thioredoxin. J. Mol. Biol. 2012, 424, 28–41. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Carreras, A.; Padilla, M.N.; Melguizo, M.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-linolenic acid is a nitric oxide donor. Nitric Oxide 2016, 57, 57–63. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. EADock: Docking of small molecules into protein active sites with a multiobjective evolutionary optimization. Proteins 2007, 67, 1010–1025. [Google Scholar] [CrossRef]
- Trott, A.; Morano, K.A. The yeast response to heat shock. In Yeast Stress Responses; Springer: Berlin/Heidelberg, Germany, 2003; pp. 71–119. [Google Scholar]
- Duina, A.A.; Chang, H.C.; Marsh, J.A.; Lindquist, S.; Gaber, R.F. A cyclophilin function in Hsp90-dependent signal transduction. Science 1996, 274, 1713–1715. [Google Scholar] [CrossRef]
- Davidson, J.F.; Schiestl, R.H. Mitochondrial respiratory electron carriers are involved in oxidative stress during heat stress in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 8483–8489. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Rand, J.D.; Grant, C.M. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell 2006, 17, 387–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weibezahn, J.; Schlieker, C.; Tessarz, P.; Mogk, A.; Bukau, B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol. Chem. 2005, 386, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Polier, S.; Dragovic, Z.; Hartl, F.U.; Bracher, A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 2008, 133, 1068–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Sun, J.; Liang, W.; Zhang, J.; Rom, O.; Garcia-Barrio, M.T.; Li, S.; Villacorta, L.; Schopfer, F.J.; Freeman, B.A.; et al. Novel gene regulatory networks identified in response to nitro-conjugated linoleic acid in human endothelial cells. Physiol. Genomics 2019, 51, 224–233. [Google Scholar] [CrossRef]
- Vollár, M.; Feigl, G.; Oláh, D.; Horváth, A.; Molnár, Á.; Kúsz, N.; Ördög, A.; Csupor, D.; Kolbert, Z. Nitro-Oleic Acid in Seeds and Differently Developed Seedlings of Brassica napus L. Plants 2020, 9, 406. [Google Scholar] [CrossRef] [Green Version]
- Balazy, M.; Poff, C.D. Biological nitration of arachidonic acid. Curr. Vasc. Pharmacol. 2004, 2, 81–93. [Google Scholar] [CrossRef]
- Baker, P.R.; Schopfer, F.J.; Sweeney, S.; Freeman, B.A. Red cell membrane and plasma linoleic acid nitration products: Synthesis, clinical identification, and quantitation. Proc. Natl. Acad. Sci. USA 2004, 101, 11577–11582. [Google Scholar] [CrossRef] [Green Version]
- Tsikas, D.; Zoerner, A.A.; Jordan, J. Oxidized and nitrated oleic acid in biological systems: Analysis by GC-MS/MS and LC-MS/MS, and biological significance. Biochim. Biophys. Acta 2011, 1811, 694–705. [Google Scholar] [CrossRef]
- Martin, C.E.; Oh, C.S.; Jiang, Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 2007, 1771, 271–285. [Google Scholar] [CrossRef]
- Tehlivets, O.; Scheuringer, K.; Kohlwein, S.D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta 2007, 1771, 255–270. [Google Scholar] [CrossRef]
- Zhao, Y.; Chang, Z.; Zhao, G.; Lu, H.; Xiong, W.; Liang, W.; Wang, H.; Villacorta, L.; Garcia-Barrio, M.T.; Zhu, T.; et al. Suppression of Vascular Macrophage Activation by Nitro-Oleic Acid and its Implication for Abdominal Aortic Aneurysm Therapy. Cardiovasc. Drugs Ther. 2021, 35, 939–951. [Google Scholar] [CrossRef]
- Pereckova, J.; Pekarova, M.; Szamecova, N.; Hoferova, Z.; Kamarytova, K.; Falk, M.; Perecko, T. Nitro-Oleic Acid Inhibits Stemness Maintenance and Enhances Neural Differentiation of Mouse Embryonic Stem Cells via STAT3 Signaling. Int. J. Mol. Sci. 2021, 22, 9981. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.L.; Heinrich, V.A.; Buchan, G.J.; O’Brien, J.P.; Uvalle, C.; Cechova, V.; Koudelka, A.; Ukani, D.; Rawas-Qalaji, M.; Oury, T.D.; et al. Nitroalkene fatty acids modulate bile acid metabolism and lung function in obese asthma. Sci. Rep. 2021, 11, 17788–17800. [Google Scholar] [CrossRef] [PubMed]
- Braumann, S.; Schumacher, W.; Im, N.G.; Nettersheim, F.S.; Mehrkens, D.; Bokredenghel, S.; Hof, A.; Nies, R.J.; Adler, C.; Winkels, H.; et al. Nitro-Oleic Acid (NO(2)-OA) Improves Systolic Function in Dilated Cardiomyopathy by Attenuating Myocardial Fibrosis. Int. J. Mol. Sci 2021, 22, 9052. [Google Scholar] [CrossRef] [PubMed]
- Biteau, B.; Labarre, J.; Toledano, M.B. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 2003, 425, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Elzahhar, P.; Belal, A.S.F.; Elamrawy, F.; Helal, N.A.; Nounou, M.I. Bioconjugation in Drug Delivery: Practical Perspectives and Future Perceptions. In Pharmaceutical Nanotechnology. Methods in Molecular Biology; Weissig, V., Elbayoumi, T., Eds.; Humana: New York, NY, USA, 2019; Volume 2000. [Google Scholar] [CrossRef]
- Rudolph, T.K.; Freeman, B.A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009, 2, re7. [Google Scholar] [CrossRef] [Green Version]
- Padilla, M.N.; Mata-Pérez, C.; Melguizo, M.; Barroso, J.B. In vitro nitro-fatty acid release from Cys-NO(2)-fatty acid adducts under nitro-oxidative conditions. Nitric Oxide 2017, 68, 14–22. [Google Scholar] [CrossRef]
- Fang, M.Y.; Huang, K.H.; Tu, W.J.; Chen, Y.T.; Pan, P.Y.; Hsiao, W.C.; Ke, Y.Y.; Tsou, L.K.; Zhang, M.M. Chemoproteomic profiling reveals cellular targets of nitro-fatty acids. Redox Biol. 2021, 46, 102126–102137. [Google Scholar] [CrossRef]








| Recombinant Tsa1 Nitroalkylated Peptides | Nitroalkylation Target |
|---|---|
| TFVcPTEIIAF | Cys 47 |
| AFIPLAFTFVcPTEIIAF | |
| AFTFVcPTEIIAF | |
| TDKNGTVLPcNW | Cys 171 |
| TDKNGTVLPcNWTPGAATIKPTVEDSKEY | |
| QWTDKNGTVLPcNW | |
| LADTNhSLSRDY | His 105 |
| ADTNhSLSRDY | |
| LADTNhSL | |
| IIDPKGVIRhITINDLPVGRNVDEAL | His 136 |
| Nitroalkylated Target | Nitroalkylation Standard: Recombinant Tsa1 Treated with NO2-OA | Endogenous Nitroalkylation (Control) | Endogenous Nitroalkylation (Heat Stress) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Nitroalkylated Peptide | m/z | RT | m/z | RT | Intensity | m/z | RT | Intensity | |
| Cys 47 | AFIPLAFTFVcPTEIIAF | 1164.65 | 117.63 | 1164.62 | 117.7 | 1.58 × 104 | ND | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aranda-Caño, L.; Valderrama, R.; Pedrajas, J.R.; Begara-Morales, J.C.; Chaki, M.; Padilla, M.N.; Melguizo, M.; López-Jaramillo, F.J.; Barroso, J.B. Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae. Antioxidants 2022, 11, 972. https://doi.org/10.3390/antiox11050972
Aranda-Caño L, Valderrama R, Pedrajas JR, Begara-Morales JC, Chaki M, Padilla MN, Melguizo M, López-Jaramillo FJ, Barroso JB. Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae. Antioxidants. 2022; 11(5):972. https://doi.org/10.3390/antiox11050972
Chicago/Turabian StyleAranda-Caño, Lorena, Raquel Valderrama, José Rafael Pedrajas, Juan C. Begara-Morales, Mounira Chaki, María N. Padilla, Manuel Melguizo, Francisco Javier López-Jaramillo, and Juan B. Barroso. 2022. "Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae" Antioxidants 11, no. 5: 972. https://doi.org/10.3390/antiox11050972
APA StyleAranda-Caño, L., Valderrama, R., Pedrajas, J. R., Begara-Morales, J. C., Chaki, M., Padilla, M. N., Melguizo, M., López-Jaramillo, F. J., & Barroso, J. B. (2022). Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae. Antioxidants, 11(5), 972. https://doi.org/10.3390/antiox11050972