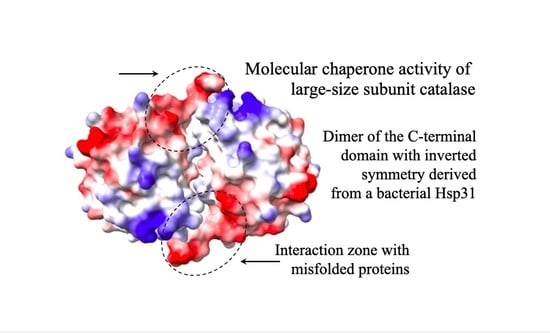
The Molecular Chaperone Mechanism of the C-Terminal Domain of Large-Size Subunit Catalases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Proteins and Reagents
2.2. Plasmid Constructs and Expression in E. coli
2.3. Enzyme Purification and Analysis
2.4. TDC Analysis and TDC3 Dimer and Monomer Formation
2.5. Chaperone Assay
2.6. Detection of Hydrophobic Regions
2.7. Unfolding Activity in the Presence of Increasing Ionic Strength or at Various pH
3. Results
3.1. Does the TDC3 Function as a Dimer or as a Monomer?
3.2. Hydrophobic Regions Are Involved in the Unfolding Activity of the CT
3.3. Charged Amino Acid Residues Are Involved in the Unfolding Activity of the CT
3.4. Identification of the Region Responsible for the Unfolding Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Finka, A.; Mattoo, R.U.; Goloubinoff, P. Experimental Milestones in the Discovery of Molecular Chaperones as Polypeptide Unfolding Enzymes. Annu. Rev. Biochem. 2016, 85, 715–742. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, S.; Hyeon, C.; Ye, X.; Lorimer, G.H.; Thirumalai, D. Molecular chaperones maximize the native state yield on biological times by driving substrates out of equilibrium. Proc. Natl. Acad. Sci. USA 2017, 114, E10919–E10927. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular Handling of Protein Aggregates by Disaggregation Machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef] [Green Version]
- Dubrez, L.; Causse, S.; Borges Bonan, N.; Dumetier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef]
- Ariosa, A.; Lee, J.H.; Wang, S.; Saraogi, I.; Shan, S.O. Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting. Proc. Natl. Acad. Sci. USA 2015, 112, E3169–E3178. [Google Scholar] [CrossRef] [Green Version]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef] [Green Version]
- Bahr, T.; Katuri, J.; Liang, T.; Bai, Y. Mitochondrial chaperones in human health and disease. Free Radic. Biol. Med. 2022, 179, 363–374. [Google Scholar] [CrossRef]
- Heldens, L.; van Genesen, S.T.; Hanssen, L.L.; Hageman, J.; Kampinga, H.H.; Lubsen, N.H. Protein refolding in peroxisomes is dependent upon an HSF1-regulated function. Cell Stress Chaperones 2012, 17, 603–613. [Google Scholar] [CrossRef] [Green Version]
- Calderwood, S.K.; Borges, T.J.; Eguchi, T.; Lang, B.J.; Murshid, A.; Okusha, Y.; Prince, T.L. Extracellular Hsp90 and protection of neuronal cells through Nrf2. Biochem. Soc. Trans. 2021, 49, 2299–2306. [Google Scholar] [CrossRef]
- Kim, H.; Wu, K.; Lee, C. Stress-Responsive Periplasmic Chaperones in Bacteria. Front. Mol. Biosci. 2021, 8, 678697. [Google Scholar] [CrossRef]
- Finka, A.; Goloubinoff, P. Proteomic data from human cell cultures refine mechanisms of chaperone-mediated protein homeostasis. Cell Stress Chaperones 2013, 18, 591–605. [Google Scholar] [CrossRef] [Green Version]
- Alagar Boopathy, L.R.; Jacob-Tomas, S.; Alecki, C.; Vera, M. Mechanisms tailoring the expression of heat shock proteins to proteostasis challenges. J. Biol. Chem. 2022, 298, 101796. [Google Scholar] [CrossRef]
- Maillot, N.J.; Honore, F.A.; Byrne, D.; Mejean, V.; Genest, O. Cold adaptation in the environmental bacterium Shewanella oneidensis is controlled by a J-domain co-chaperone protein network. Commun. Biol. 2019, 2, 323. [Google Scholar] [CrossRef] [Green Version]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef]
- Koldewey, P.; Horowitz, S.; Bardwell, J.C.A. Chaperone-client interactions: Non-specificity engenders multifunctionality. J. Biol. Chem. 2017, 292, 12010–12017. [Google Scholar] [CrossRef] [Green Version]
- Rizzolo, K.; Houry, W.A. Multiple functionalities of molecular chaperones revealed through systematic mapping of their interaction networks. J. Biol. Chem. 2019, 294, 2142–2150. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.J.; Kim, S.; Kim, Y.J.; Kim, M.K.; Kang, S.G.; Lee, J.H.; Kim, W.; Cha, S.S. Dissection of the dimerization modes in the DJ-1 superfamily. Mol. Cells 2012, 33, 163–171. [Google Scholar] [CrossRef]
- Mogk, A.; Ruger-Herreros, C.; Bukau, B. Cellular Functions and Mechanisms of Action of Small Heat Shock Proteins. Annu. Rev. Microbiol. 2019, 73, 89–110. [Google Scholar] [CrossRef]
- Libich, D.S.; Tugarinov, V.; Clore, G.M. Intrinsic unfoldase/foldase activity of the chaperonin GroEL directly demonstrated using multinuclear relaxation-based NMR. Proc. Natl. Acad. Sci. USA 2015, 112, 8817–8823. [Google Scholar] [CrossRef] [Green Version]
- Salmon, L.; Ahlstrom, L.S.; Horowitz, S.; Dickson, A.; Brooks, C.L., 3rd; Bardwell, J.C. Capturing a Dynamic Chaperone-Substrate Interaction Using NMR-Informed Molecular Modeling. J. Am. Chem. Soc. 2016, 138, 9826–9839. [Google Scholar] [CrossRef] [Green Version]
- Klotz, M.G.; Loewen, P.C. The molecular evolution of catalatic hydroperoxidases: Evidence for multiple lateral transfer of genes between prokaryota and from bacteria into eukaryota. Mol. Biol. Evol. 2003, 20, 1098–1112. [Google Scholar] [CrossRef]
- Zamocky, M.; Gasselhuber, B.; Furtmuller, P.G.; Obinger, C. Molecular evolution of hydrogen peroxide degrading enzymes. Arch. Biochem. Biophys 2012, 525, 131–144. [Google Scholar] [CrossRef] [Green Version]
- Hansberg, W.; Salas-Lizana, R.; Dominguez, L. Fungal catalases: Function, phylogenetic origin and structure. Arch. Biochem. Biophys 2012, 525, 170–180. [Google Scholar] [CrossRef]
- Hansberg, W. Monofunctional Heme-Catalases. Antioxidants 2022, 11, 2173. [Google Scholar] [CrossRef]
- Hansberg, W.; Nava-Ramirez, T.; Rangel-Silva, P.; Diaz-Vilchis, A.; Mendoza-Oliva, A. Large-Size Subunit Catalases Are Chimeric Proteins: A H(2)O(2) Selecting Domain with Catalase Activity Fused to a Hsp31-Derived Domain Conferring Protein Stability and Chaperone Activity. Antioxidants 2022, 11, 979. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Cookson, M.R. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 2004, 4, 6. [Google Scholar] [CrossRef] [Green Version]
- Horvath, M.M.; Grishin, N.V. The C-terminal domain of HPII catalase is a member of the type I glutamine amidotransferase superfamily. Proteins 2001, 42, 230–236. [Google Scholar] [CrossRef]
- Hansberg, W.; Aguirre, J. Hyperoxidant states cause microbial cell differentiation by cell isolation from dioxygen. J. Theor. Biol. 1990, 142, 201–221. [Google Scholar] [CrossRef]
- Aguirre, J.; Rios-Momberg, M.; Hewitt, D.; Hansberg, W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005, 13, 111–118. [Google Scholar] [CrossRef]
- Hansberg, W.; Aguirre, J.; Rís-Momberg, M.; Rangel, P.; Peraza, L.; de Oca, Y.M.; Cano-Domínguez, N. Cell differentiation as a response to oxidative stress. In Stress in Yeasts and Filamentous Fungi; Avery, S.V., Stratford, M., West, P.V., Eds.; Elsevier Limited: London, UK, 2008; Volume 27, pp. 235–257. [Google Scholar]
- Michan, S.; Lledias, F.; Baldwin, J.D.; Natvig, D.O.; Hansberg, W. Regulation and oxidation of two large monofunctional catalases. Free. Radic. Biol. Med. 2002, 33, 521–532. [Google Scholar] [CrossRef]
- Diaz, A.; Rangel, P.; Montes de Oca, Y.; Lledias, F.; Hansberg, W. Molecular and kinetic study of catalase-1, a durable large catalase of Neurospora crassa. Free. Radic. Biol. Med. 2001, 31, 1323–1333. [Google Scholar] [CrossRef]
- Nava-Ramirez, T.; Hansberg, W. Chaperone activity of large-size subunit catalases. Free. Radic. Biol. Med. 2020, 156, 99–106. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Pescitelli, R.; Camici, G.; Manao, G.; Ramponi, G. Hydrogen peroxide triggers the formation of a disulfide dimer of muscle acylphosphatase and modifies some functional properties of the enzyme. J. Biol. Chem. 2001, 276, 41862–41869. [Google Scholar] [CrossRef] [Green Version]
- Gorin, G.; Martic, P.A.; Doughty, G. Kinetics of the reaction of N-ethylmaleimide with cysteine and some congeners. Arch. Biochem. Biophys 1966, 115, 593–597. [Google Scholar] [CrossRef]
- Hill, B.G.; Ramana, K.V.; Cai, J.; Bhatnagar, A.; Srivastava, S.K. Measurement and identification of S-glutathiolated proteins. Methods Enzymol. 2010, 473, 179–197. [Google Scholar] [CrossRef] [Green Version]
- Koldewey, P.; Stull, F.; Horowitz, S.; Martin, R.; Bardwell, J.C.A. Forces Driving Chaperone Action. Cell 2016, 166, 369–379. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Choi, D.; Cha, S.Y.; Oh, Y.M.; Hwang, E.; Park, C.; Ryu, K.S. Zinc-mediated Reversible Multimerization of Hsp31 Enhances the Activity of Holding Chaperone. J. Mol. Biol. 2018, 430, 1760–1772. [Google Scholar] [CrossRef]
- Diaz, A.; Valdes, V.J.; Rudino-Pinera, E.; Horjales, E.; Hansberg, W. Structure-function relationships in fungal large-subunit catalases. J. Mol. Biol. 2009, 386, 218–232. [Google Scholar] [CrossRef]
- Sastry, M.S.; Korotkov, K.; Brodsky, Y.; Baneyx, F. Hsp31, the Escherichia coli yedU gene product, is a molecular chaperone whose activity is inhibited by ATP at high temperatures. J. Biol. Chem. 2002, 277, 46026–46034. [Google Scholar] [CrossRef] [Green Version]
- Negi, S.S.; Schein, C.H.; Oezguen, N.; Power, T.D.; Braun, W. InterProSurf: A web server for predicting interacting sites on protein surfaces. Bioinformatics 2007, 23, 3397–3399. [Google Scholar] [CrossRef]
- Pires, D.E.; Ascher, D.B.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Castelao, B.; Munoz, C.; Sanchez, I.; Goethals, M.; Vandekerckhove, J.; Castano, J.G. Reduced protein stability of human DJ-1/PARK7 L166P, linked to autosomal recessive Parkinson disease, is due to direct endoproteolytic cleavage by the proteasome. Biochim. Biophys Acta. 2012, 1823, 524–533. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Wang, G.; Cha, J.Y.; Li, G.; Chen, S.; Li, Z.; Guo, J.; Zhang, C.; Yang, Y.; et al. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 2015, 27, 908–925. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Cui, L.; Xie, Z.; Zhang, Z.; Liu, E.; Peng, X. Two NCA1 isoforms interact with catalase in a mutually exclusive manner to redundantly regulate its activity in rice. BMC Plant Biol. 2019, 19, 105. [Google Scholar] [CrossRef]
- Li, G.; Li, J.; Hao, R.; Guo, Y. Activation of catalase activity by a peroxisome-localized small heat shock protein Hsp17.6CII. J. Genet. Genom. 2017, 44, 395–404. [Google Scholar] [CrossRef]
- Nisaa, K.; Ben-Zvi, A. Chaperone networks are shaped by cellular differentiation and identity. Trends Cell Biol. 2022, 32, 470–474. [Google Scholar] [CrossRef]
- Rhee, S.G.; Woo, H.A.; Kang, D. The Role of Peroxiredoxins in the Transduction of H(2)O(2) Signals. Antioxid. Redox Signal. 2018, 28, 537–557. [Google Scholar] [CrossRef]
- Berndt, C.; Lillig, C.H.; Holmgren, A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim. Biophys. Acta 2008, 1783, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, X.; Wang, C.C. Protein disulfide-isomerase, a folding catalyst and a redox-regulated chaperone. Free. Radic. Biol. Med. 2015, 83, 305–313. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nava-Ramírez, T.; Gutiérrez-Terrazas, S.; Hansberg, W. The Molecular Chaperone Mechanism of the C-Terminal Domain of Large-Size Subunit Catalases. Antioxidants 2023, 12, 839. https://doi.org/10.3390/antiox12040839
Nava-Ramírez T, Gutiérrez-Terrazas S, Hansberg W. The Molecular Chaperone Mechanism of the C-Terminal Domain of Large-Size Subunit Catalases. Antioxidants. 2023; 12(4):839. https://doi.org/10.3390/antiox12040839
Chicago/Turabian StyleNava-Ramírez, Teresa, Sammy Gutiérrez-Terrazas, and Wilhelm Hansberg. 2023. "The Molecular Chaperone Mechanism of the C-Terminal Domain of Large-Size Subunit Catalases" Antioxidants 12, no. 4: 839. https://doi.org/10.3390/antiox12040839
APA StyleNava-Ramírez, T., Gutiérrez-Terrazas, S., & Hansberg, W. (2023). The Molecular Chaperone Mechanism of the C-Terminal Domain of Large-Size Subunit Catalases. Antioxidants, 12(4), 839. https://doi.org/10.3390/antiox12040839