Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses
Abstract
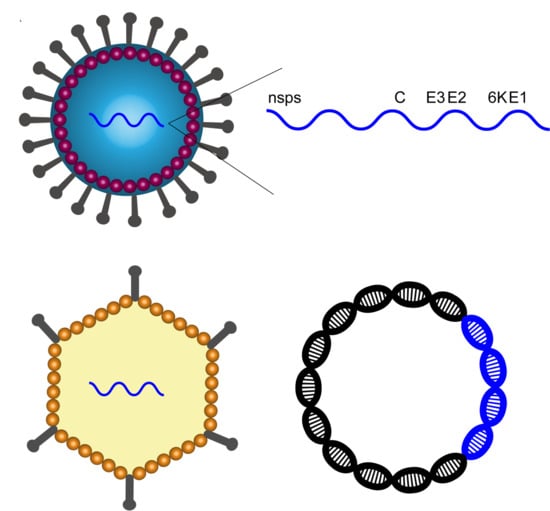
:1. Introduction
2. Animal Models and Strain Selection for Vaccine Evaluation
2.1. Animal Models
2.2. Viral Strain Selection
3. Vaccine Strategies
4. Recent Progress in the Development of Viral Vector Based Vaccines
4.1. Adenovirus Vector
4.2. Eilat Virus Vector
4.3. Equine Herpesvirus Vector
4.4. Isfahan Virus Vector
4.5. Sindbis Virus Vector
4.6. Vaccinia Virus Vector
4.7. Vesicular Stomatitis Virus Vector
5. Recent Progress in the Development of Plasmid DNA Vaccines
5.1. DNA Vaccines for EEEV
5.2. DNA Vaccines for VEEV
5.3. DNA Vaccines for WEEV
5.4. Trivalent DNA Vaccines
6. Recent Progress in the Development of RNA Vaccines
7. Evaluation of Candidate Vaccine Sequences
8. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weaver, S.C.; Forrester, N.L. Chikungunya: Evolutionary history and recent epidemic spread. Antivir. Res. 2015, 120, 32–39. [Google Scholar] [CrossRef]
- Pettersson, J.H.-O.; Eldholm, V.; Seligman, S.J.; Lundkvist, Å.; Falconar, A.K.; Gaunt, M.W.; Musso, D.; Nougairède, A.; Charrel, R.; Gould, E.A. How did Zika virus emerge in the Pacific Islands and Latin America? MBio 2016, 7, e01239-16. [Google Scholar] [CrossRef] [Green Version]
- Zumla, A.; Dar, O.; Kock, R.; Muturi, M.; Ntoumi, F.; Kaleebu, P.; Eusebio, M.; Mfinanga, S.; Bates, M.; Mwaba, P.; et al. Taking forward a ‘One Health’ approach for turning the tide against the Middle East respiratory syndrome coronavirus and other zoonotic pathogens with epidemic potential. Int. J. Infect. Dis. 2016, 47, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.H.; Strych, U.; Hotez, P.J.; Bottazzi, M.E. The SARS-CoV-2 vaccine pipeline: An overview. Curr. Trop. Med. Rep. 2020, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Saif, L.J. Vaccines for COVID-19: Perspectives, prospects, and challenges based on candidate SARS, MERS, and animal coronavirus vaccines. Eur. Med. J. 2020. [Google Scholar] [CrossRef] [Green Version]
- Peeri, N.C.; Shrestha, N.; Rahman, M.S.; Zaki, R.; Tan, Z.; Bibi, S.; Baghbanzadeh, M.; Aghamohammadi, N.; Zhang, W.; Haque, U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: What lessons have we learned? Int. J. Epidemiol. 2020, dyaa033, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Gould, E.; Pettersson, J.; Higgs, S.; Charrel, R.; de Lamballerie, X. Emerging arboviruses: Why today? One Health 2017, 4, 1–13. [Google Scholar] [CrossRef]
- Bloom, D.E.; Black, S.; Rappuoli, R. Emerging infectious diseases: A proactive approach. Proc. Natl. Acad. Sci. USA 2017, 114, 4055–4059. [Google Scholar] [CrossRef] [Green Version]
- Forrester, N.L.; Wertheim, J.O.; Dugan, V.G.; Auguste, A.J.; Lin, D.; Adams, A.P.; Chen, R.; Gorchakov, R.; Leal, G.; Estrada-Franco, J.G. Evolution and spread of Venezuelan equine encephalitis complex Alphavirus in the Americas. PLoS Negl. Trop. Dis. 2017, 11, e0005693. [Google Scholar] [CrossRef]
- Ronca, S.E.; Dineley, K.T.; Paessler, S. Neurological sequelae resulting from encephalitic Alphavirus infection. Front. Microbiol. 2016, 7, 959. [Google Scholar] [CrossRef] [PubMed]
- Villari, P.; Spielman, A.; Komar, N.; McDowell, M.; Timperi, R.J. The economic burden imposed by a residual case of Eastern encephalitis. Am. J. Trop. Med. Hyg. 1995, 52, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Broeck, G.T.; Merrell, M.H. A serological difference between eastern and western equine encephalomyelitis virus. Proc. Soc. Exp. Biol. Med. 1933, 31, 217–220. [Google Scholar] [CrossRef]
- Carrera, J.P.; Forrester, N.; Wang, E.; Vittor, A.Y.; Haddow, A.D.; Lopez-Verges, S.; Abadia, I.; Castano, E.; Sosa, N.; Baez, C.; et al. Eastern equine encephalitis in Latin America. N. Engl. J. Med. 2013, 369, 732–744. [Google Scholar] [CrossRef] [Green Version]
- Kubes, V.; Rios, F.A. The causative agent of infectious equine encephalomyelitis in Venezuela. Science 1939, 90, 20–21. [Google Scholar] [CrossRef]
- Suarez, O.M.; Bergold, G.H. Investigations of an outbreak of Venezuelan equine encephalitis in towns of eastern Venezuela. Am. J. Trop. Med. Hyg. 1968, 17, 875–880. [Google Scholar] [CrossRef]
- Bowen, G.S.; Fashinell, T.R.; Dean, P.B.; Gregg, M.B. Clinical aspects of human Venezuelan equine encephalitis in Texas. Bull. Pan Am. Health Organ. 1976, 10, 46–57. [Google Scholar]
- Rossi, A.L. Rural epidemic encephalitis in Venezuela caused by a group A arbovirus (VEE). Prog. Med. Virol. 1967, 9, 176–203. [Google Scholar]
- Weaver, S.C.; Ferro, C.; Barrera, R.; Boshell, J.; Navarro, J.C. Venezuelan equine encephalitis. Annu. Rev. Entomol. 2004, 49, 141–174. [Google Scholar] [CrossRef]
- Meyer, K.F.; Haring, C.M.; Howitt, B. The etiology of epizootic encephalomyelitis of horses in the San Joaquin Valley, 1930. Science 1931, 74, 227–228. [Google Scholar] [CrossRef]
- Arechiga-Ceballos, N.; Aguilar-Setien, A. Alphaviral equine encephalomyelitis (Eastern, Western and Venezuelan). Rev. Sci. Tech. Off. Int. Epiz. 2015, 34, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.; Reed, D.; Hutt, J. Aerobiology and inhalation exposure to biological select agents and toxins. Vet. Pathol. 2010, 47, 779–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, D.N.; Heppner, D.G.; Gardner, S.N.; Jaing, C.; Dupuy, L.C.; Schmaljohn, C.S.; Carlton, K. Current strategic thinking for the development of a trivalent Alphavirus vaccine for human use. Am. J. Trop. Med. Hyg. 2014, 91, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, N.C.; Adams, A.P.; Weaver, S.C. Evolutionary patterns of Eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J. Virol. 2010, 84, 1014–1025. [Google Scholar] [CrossRef] [Green Version]
- Zacks, M.A.; Paessler, S. Encephalitic alphaviruses. Vet. Microbiol. 2010, 140, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.; Wan, J.; Kielian, M. The Alphavirus exit pathway: What we know and what we wish we knew. Viruses 2018, 10, 89. [Google Scholar] [CrossRef] [Green Version]
- Olivia, L.W.; Obanda, V.; Bucht, G.; Mosomtai, G.; Otieno, V.; Ahlm, C.; Evander, M. Global emergence of Alphaviruses that cause arthritis in humans. Infect. Ecol. Epidemiol. 2015, 5, 29853. [Google Scholar] [CrossRef] [Green Version]
- Weaver, S.C.; Paessler, S. Chapter 21—Alphaviral encephalitides. In Vaccines for Biodefense and Emerging and Neglected Diseases; Barrett, A.D.T., Stanberry, L.R., Eds.; Academic Press: Amsterdam, The Netherlands, 2009; pp. 339–359. [Google Scholar]
- Cox, H.R.; Olitsky, P.K. Prevention of experimental equine encephalomyelitis in guinea pigs by means of virus adsorbed on aluminum hydroxide. Science 1934, 79, 459. [Google Scholar] [CrossRef]
- Beard, J.; Finkelstein, H.; Sealy, W.; Wyckoff, R. Immunization against equine encephalomyelitis with chick embryo vaccines. Science 1938, 87, 490. [Google Scholar] [CrossRef]
- Pittman, P.R.; Makuch, R.S.; Mangiafico, J.A.; Cannon, T.L.; Gibbs, P.H.; Peters, C.J. Long-term duration of detectable neutralizing antibodies after administration of live-attenuated VEE vaccine and following booster vaccination with inactivated VEE vaccine. Vaccine 1996, 14, 337–343. [Google Scholar] [CrossRef]
- Cieslak, T.J.; Christopher, G.W.; Kortepeter, M.G.; Rowe, J.R.; Pavlin, J.A.; Culpepper, R.C.; Eitzen, E.M., Jr. Immunization against potential biological warfare agents. Clin. Infect. Dis. 2000, 30, 843–850. [Google Scholar] [CrossRef]
- Berge, T.O.; Banks, I.S.; Tigertt, W. Attenuation of Venezuelan equine encephalomyelitis virus by ire vitro cultivation in guinea-pig heart cells. Am. J. Hyg. 1961, 73, 209–218. [Google Scholar]
- Kinney, R.M.; Johnson, B.J.; Welch, J.B.; Tsuchiya, K.R.; Trent, D.W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 1989, 170, 19–30. [Google Scholar] [CrossRef]
- Kinney, R.M.; Chang, G.; Tsuchiya, K.R.; Sneider, J.M.; Roehrig, J.; Woodward, T.; Trent, D. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5’-noncoding region and the E2 envelope glycoprotein. J. Virol. 1993, 67, 1269–1277. [Google Scholar] [CrossRef] [Green Version]
- Rayfield, E.; Gorelkin, L.; Cumow, R.; Jahrling, P. Virus-induced pancreatid disease induced by Venezuelan equine encephalitis virus. Alterations in glucose tolerance and insulin release. J. Am. Diab. Assoc. 1976, 25, 621–623. [Google Scholar]
- Casamassima, A.C.; Hess, L.W.; Marty, A. TC-83 Venezuelan equine encephalitis vaccine exposure during pregnancy. Teratology 1987, 36, 287–289. [Google Scholar] [CrossRef]
- Jahrling, P.B.; Stephenson, E.H. Protective efficacies of live attenuated and formaldehyde-inactivated Venezuelan equine encephalitis virus vaccines against aerosol challenge in hamsters. J. Clin. Microbiol. 1984, 19, 429–431. [Google Scholar] [CrossRef] [Green Version]
- Edelman, R.; Ascher, M.S.; Oster, C.N.; Ramsburg, H.H.; Cole, F.E.; Eddy, G.A. Evaluation in humans of a new, inactivated vaccine for Venezuelan equine encephalitis virus (C-84). J. Infect. Dis. 1979, 140, 708–715. [Google Scholar] [CrossRef]
- Kinney, R.M.; Tsuchiya, K.R.; Sneider, J.M.; Trent, D.W. Molecular evidence for the origin of the widespread Venezuelan equine encephalitis epizootic of 1969 to 1972. J. Gen. Virol. 1992, 73, 3301–3305. [Google Scholar] [CrossRef]
- Reisler, R.B.; Gibbs, P.H.; Danner, D.K.; Boudreau, E.F. Immune interference in the setting of same-day administration of two similar inactivated Alphavirus vaccines: Eastern equine and Western equine encephalitis. Vaccine 2012, 30, 7271–7277. [Google Scholar] [CrossRef]
- Pittman, P.R.; Liu, C.T.; Cannon, T.L.; Mangiafico, J.A.; Gibbs, P.H. Immune interference after sequential Alphavirus vaccine vaccinations. Vaccine 2009, 27, 4879–4882. [Google Scholar] [CrossRef] [PubMed]
- Steele, K.; Twenhafel, N. Pathology of animal models of Alphavirus encephalitis. Vet. Pathol. 2010, 47, 790–805. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.C.; SenGupta, S.K.; Smith, J.F. Pathogenesis of Venezuelan equine encephalitis virus infection in mice and hamsters. Vet. Pathol. 1991, 28, 410–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudley, D.M.; Newman, C.M.; Lalli, J.; Stewart, L.M.; Koenig, M.R.; Weiler, A.M.; Semler, M.R.; Barry, G.L.; Zarbock, K.R.; Mohns, M.S. Infection via mosquito bite alters Zika virus tissue tropism and replication kinetics in rhesus macaques. Nat. Commun. 2017, 8, 2096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusnak, J.M.; Glass, P.J.; Weaver, S.C.; Sabourin, C.L.; Glenn, A.M.; Klimstra, W.; Badorrek, C.S.; Nasar, F.; Ward, L.A. Approach to strain selection and the propagation of viral stocks for Venezuelan equine encephalitis virus vaccine efficacy testing under the animal rule. Viruses 2019, 11, 807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Höper, D.; Freuling, C.M.; Müller, T.; Hanke, D.; von Messling, V.; Duchow, K.; Beer, M.; Mettenleiter, T.C. High definition viral vaccine strain identity and stability testing using full-genome population data—The next generation of vaccine quality control. Vaccine 2015, 33, 5829–5837. [Google Scholar] [CrossRef]
- Sharma, A.; Knollmann-Ritschel, B. Current understanding of the molecular basis of Venezuelan equine encephalitis virus pathogenesis and vaccine development. Viruses 2019, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Turell, M.J.; Ludwig, G.V.; Kondig, J.; Smith, J.F. Limited potential for mosquito transmission of genetically engineered, live-attenuated Venezuelan equine encephalitis virus vaccine candidates. Am. J. Trop. Med. Hyg. 1999, 60, 1041–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedersen, C.E., Jr.; Robinson, D.M.; Cole, F.E., Jr. Isolation of the vaccine strain of Venezuelan equine encephalomyelitis virus from mosquitoes in Louisiana. Am. J. Epidemiol. 1972, 95, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Papamichail, D.; Coleman, J.R.; Skiena, S.; Wimmer, E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 2006, 80, 9687–9696. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Skiena, S.; Futcher, B.; Mueller, S.; Wimmer, E. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 9481–9486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.Y.; Ortiz-Riano, E.; Nogales, A.; de la Torre, J.C.; Martinez-Sobrido, L. Development of live-attenuated arenavirus vaccines based on codon deoptimization. J. Virol. 2015, 89, 3523–3533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouen, C.L.; Collins, P.L.; Buchholz, U.J. Attenuation of human respiratory viruses by synonymous genome recoding. Front. Immunol. 2019, 10, 1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bull, J.J. Evolutionary reversion of live viral vaccines: Can genetic engineering subdue it? Virus Evol. 2015, 1, vev005. [Google Scholar] [CrossRef]
- Lauring, A.S.; Jones, J.O.; Andino, R. Rationalizing the development of live attenuated virus vaccines. Nat. Biotechnol. 2010, 28, 573–579. [Google Scholar] [CrossRef]
- Pushko, P.; Lukashevich, I.S.; Weaver, S.C.; Tretyakova, I. DNA-launched live-attenuated vaccines for biodefense applications. Expert Rev. Vaccines 2016, 15, 1223–1234. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.M.; Sokoloski, K.J.; Jokinen, J.D.; Pfeffer, T.L.; Chu, Y.K.; Adcock, R.S.; Chung, D.; Tretyakova, I.; Pushko, P.; Lukashevich, I.S. Advanced safety and genetic stability in mice of a novel DNA-launched Venezuelan equine encephalitis virus vaccine with rearranged structural genes. Vaccines 2020, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Tretyakova, I.; Tibbens, A.; Jokinen, J.D.; Johnson, D.M.; Lukashevich, I.S.; Pushko, P. Novel DNA-launched Venezuelan equine encephalitis virus vaccine with rearranged genome. Vaccine 2019, 37, 3317–3325. [Google Scholar] [CrossRef]
- Phillpotts, R.J. Venezuelan equine encephalitis virus complex-specific monoclonal antibody provides broad protection, in murine models, against airborne challenge with viruses from serogroups I, II and III. Virus Res. 2006, 120, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.D.; Buckley, M.J.; Melanson, V.R.; Glass, P.J.; Norwood, D.; Hart, M.K. Antibody to the E3 glycoprotein protects mice against lethal Venezuelan equine encephalitis virus infection. J. Virol. 2010, 84, 12683–12690. [Google Scholar] [CrossRef] [Green Version]
- Reed, D.S.; Glass, P.J.; Bakken, R.R.; Barth, J.F.; Lind, C.M.; da Silva, L.; Hart, M.K.; Rayner, J.; Alterson, K.; Custer, M.; et al. Combined Alphavirus replicon particle vaccine induces durable and cross-protective immune responses against equine encephalitis viruses. J. Virol. 2014, 88, 12077–12086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saxena, M.; Van, T.T.H.; Baird, F.J.; Coloe, P.J.; Smooker, P.M. Pre-existing immunity against vaccine vectors–friend or foe? Microbiology 2013, 159, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, G.L.; Mackett, M.; Moss, B. Infectious vaccinia virus recombinants that express hepatitis B virus surface antigen. Nature 1983, 302, 490. [Google Scholar] [CrossRef] [PubMed]
- Draper, S.J.; Heeney, J.L. Viruses as vaccine vectors for infectious diseases and cancer. Nat. Rev. Microbiol. 2010, 8, 62. [Google Scholar] [CrossRef]
- Price, G.E.; Soboleski, M.R.; Lo, C.-Y.; Misplon, J.A.; Quirion, M.R.; Houser, K.V.; Pearce, M.B.; Pappas, C.; Tumpey, T.M.; Epstein, S.L. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS ONE 2010, 5, e13162. [Google Scholar] [CrossRef] [Green Version]
- Phillpotts, R.; O’brien, L.; Appleton, R.; Carr, S.; Bennett, A. Intranasal immunisation with defective adenovirus serotype 5 expressing the Venezuelan equine encephalitis virus E2 glycoprotein protects against airborne challenge with virulent virus. Vaccine 2005, 23, 1615–1623. [Google Scholar] [CrossRef]
- Perkins, S.D.; Williams, A.J.; O’Brien, L.M.; Laws, T.R.; Phillpotts, R.J. CpG used as an adjuvant for an adenovirus-based Venezuelan equine encephalitis virus vaccine increases the immune response to the vector, but not to the transgene product. Viral Immunol. 2008, 21, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.J.; O’Brien, L.M.; Phillpotts, R.J.; Perkins, S.D. Improved efficacy of a gene optimised adenovirus-based vaccine for Venezuelan equine encephalitis virus. Virol. J. 2009, 6, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.Q.; Barabé, N.D.; Chau, D.; Wong, C.; Rayner, G.R.; Hu, W.-G.; Nagata, L.P. Complete protection of mice against a lethal dose challenge of Western equine encephalitis virus after immunization with an adenovirus-vectored vaccine. Vaccine 2007, 25, 4368–4375. [Google Scholar] [CrossRef]
- Barabé, N.D.; Rayner, G.A.; Christopher, M.E.; Nagata, L.P.; Wu, J.Q. Single-dose, fast-acting vaccine candidate against Western equine encephalitis virus completely protects mice from intranasal challenge with different strains of the virus. Vaccine 2007, 25, 6271–6276. [Google Scholar] [CrossRef]
- Swayze, R.D.; Bhogal, H.S.; Barabé, N.D.; McLaws, L.J.; Wu, J.Q. Envelope protein E1 as vaccine target for Western equine encephalitis virus. Vaccine 2011, 29, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Seymour, R.L.; Kaelber, J.T.; Kim, D.Y.; Leal, G.; Sherman, M.B.; Frolov, I.; Chiu, W.; Weaver, S.C.; Nasar, F. Novel insect-specific Eilat virus-based chimeric vaccine candidates provide durable, mono-and multivalent, single-dose protection against lethal Alphavirus challenge. J. Virol. 2018, 92, e01274-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosas, C.T.; Paessler, S.; Ni, H.; Osterrieder, N. Protection of mice by equine herpesvirus type 1–based experimental vaccine against lethal Venezuelan equine encephalitis virus infection in the absence of neutralizing antibodies. Am. J. Trop. Med. Hyg. 2008, 78, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Nasar, F.; Matassov, D.; Seymour, R.L.; Latham, T.; Gorchakov, R.V.; Nowak, R.M.; Leal, G.; Hamm, S.; Eldridge, J.H.; Tesh, R.B. Recombinant Isfahan virus and vesicular stomatitis virus vaccine vectors provide durable, multivalent, single dose protection against lethal Alphavirus challenge. J. Virol. 2017, 91, e01729-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, E.; Petrakova, O.; Adams, A.P.; Aguilar, P.V.; Kang, W.; Paessler, S.; Volk, S.M.; Frolov, I.; Weaver, S.C. Chimeric Sindbis/Eastern equine encephalitis vaccine candidates are highly attenuated and immunogenic in mice. Vaccine 2007, 25, 7573–7581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, C.J.; Adams, A.P.; Wang, E.; Leal, G.; Seymour, R.L.; Sivasubramani, S.K.; Mega, W.; Frolov, I.; Didier, P.J.; Weaver, S.C. A chimeric Sindbis-based vaccine protects cynomolgus macaques against a lethal aerosol challenge of Eastern equine encephalitis virus. Vaccine 2013, 31, 1464–1470. [Google Scholar] [CrossRef] [Green Version]
- Paessler, S.; Fayzulin, R.Z.; Anishchenko, M.; Greene, I.P.; Weaver, S.C.; Frolov, I. Recombinant Sindbis/Venezuelan equine encephalitis virus is highly attenuated and immunogenic. J. Virol. 2003, 77, 9278–9286. [Google Scholar] [CrossRef] [Green Version]
- Paessler, S.; Ni, H.; Petrakova, O.; Fayzulin, R.Z.; Yun, N.; Anishchenko, M.; Weaver, S.C.; Frolov, I. Replication and clearance of Venezuelan equine encephalitis virus from the brains of animals vaccinated with chimeric SIN/VEE viruses. J. Virol. 2006, 80, 2784–2796. [Google Scholar] [CrossRef] [Green Version]
- Atasheva, S.; Wang, E.; Adams, A.P.; Plante, K.S.; Ni, S.; Taylor, K.; Miller, M.E.; Frolov, I.; Weaver, S.C. Chimeric Alphavirus vaccine candidates protect mice from intranasal challenge with Western equine encephalitis virus. Vaccine 2009, 27, 4309–4319. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.-G.; Steigerwald, R.; Kalla, M.; Volkmann, A.; Noll, D.; Nagata, L.P. Protective efficacy of monovalent and trivalent recombinant MVA-based vaccines against three encephalitic alphaviruses. Vaccine 2018, 36, 5194–5203. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, D. Adenoviral vector-based strategies against infectious disease and cancer. Hum. Vaccines Immunother. 2016, 12, 2064–2074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundstrom, K. Alphavirus-based vaccines. Viruses 2014, 6, 2392–2415. [Google Scholar] [CrossRef] [Green Version]
- Nasar, F.; Palacios, G.; Gorchakov, R.V.; Guzman, H.; Da Rosa, A.P.T.; Savji, N.; Popov, V.L.; Sherman, M.B.; Lipkin, W.I.; Tesh, R.B. Eilat virus, a unique Alphavirus with host range restricted to insects by RNA replication. Proc. Natl. Acad. Sci. USA 2012, 109, 14622–14627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolling, B.; Weaver, S.; Tesh, R.; Vasilakis, N. Insect-specific virus discovery: Significance for the arbovirus community. Viruses 2015, 7, 4911–4928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.-X.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Kang, Y.-J.; Chen, L.-J.; Qin, X.-C.; Xu, J.; Holmes, E.C.; Zhang, Y.-Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 2015, 4, e05378. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Auguste, A.J.; Kaelber, J.T.; Luo, H.; Rossi, S.L.; Fenton, K.; Leal, G.; Kim, D.Y.; Chiu, W.; Wang, T. A Chikungunya fever vaccine utilizing an insect-specific virus platform. Nat. Med. 2017, 23, 192. [Google Scholar] [CrossRef]
- Trapp, S.; von Einem, J.; Hofmann, H.; Köstler, J.; Wild, J.; Wagner, R.; Beer, M.; Osterrieder, N. Potential of equine herpesvirus 1 as a vector for immunization. J. Virol. 2005, 79, 5445–5454. [Google Scholar] [CrossRef] [Green Version]
- Tesh, R.; Saidi, S.; Javadian, E.; Loh, P.; Nadim, A. Isfahan virus, a new vesiculovirus infecting humans, gerbils, and sandflies in Iran. Am. J. Trop. Med. Hyg. 1977, 26, 299–306. [Google Scholar] [CrossRef]
- Adouchief, S.; Smura, T.; Sane, J.; Vapalahti, O.; Kurkela, S. Sindbis virus as a human pathogen—Epidemiology, clinical picture and pathogenesis. Rev. Med. Virol. 2016, 26, 221–241. [Google Scholar] [CrossRef]
- Overton, E.T.; Lawrence, S.J.; Wagner, E.; Nopora, K.; Rösch, S.; Young, P.; Schmidt, D.; Kreusel, C.; De Carli, S.; Meyer, T.P. Immunogenicity and safety of three consecutive production lots of the non replicating smallpox vaccine MVA: A randomised, double blind, placebo controlled phase III trial. PLoS ONE 2018, 13, e0195897. [Google Scholar] [CrossRef] [Green Version]
- Tober, R.; Banki, Z.; Egerer, L.; Muik, A.; Behmüller, S.; Kreppel, F.; Greczmiel, U.; Oxenius, A.; von Laer, D.; Kimpel, J. VSV-GP: A potent viral vaccine vector that boosts the immune response upon repeated applications. J. Virol. 2014, 88, 4897–4907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisbert, T.W.; Feldmann, H. Recombinant vesicular stomatitis virus–based vaccines against Ebola and Marburg virus infections. J. Infect. Dis. 2011, 204, S1075–S1081. [Google Scholar] [CrossRef] [PubMed]
- Emanuel, J.; Callison, J.; Dowd, K.A.; Pierson, T.C.; Feldmann, H.; Marzi, A. A VSV-based Zika virus vaccine protects mice from lethal challenge. Sci. Rep. 2018, 8, 11043. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kumar, S.A.; Jhan, Y.Y.; Bishop, C.J. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018, 80, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhang, F.; Xu, S.; Fire, A.Z.; Kay, M.A. The extragenic spacer length between the 5′ and 3’ ends of the transgene expression cassette affects transgene silencing from plasmid-based vectors. Mol. Ther. 2012, 20, 2111–2119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, J.; Rodriguez, S.; Finlayson, N.; Williams, J.; Carnes, A. Antibiotic-free production of a herpes simplex virus 2 DNA vaccine in a high yield cGMP process. Hum. Vaccines Immunother. 2013, 9, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Suschak, J.J.; Williams, J.A.; Schmaljohn, C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccines Immunother. 2017, 13, 2837–2848. [Google Scholar] [CrossRef] [Green Version]
- Garmory, H.S.; Brown, K.A.; Titball, R.W. DNA vaccines: Improving expression of antigens. Genet. Vaccines Ther. 2003, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.H.; Suh, Y.S.; Sung, J.H.; Yang, S.H.; Sung, Y.C. Comparison of various expression plasmids for the induction of immune response by DNA immunization. Mol. Cells 1997, 7, 495–501. [Google Scholar]
- Faurez, F.; Dory, D.; Le Moigne, V.; Gravier, R.; Jestin, A. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine 2010, 28, 3888–3895. [Google Scholar] [CrossRef] [Green Version]
- Ljungberg, K.; Liljestrom, P. Self-replicating alphavirus RNA vaccines. Expert Rev. Vaccines 2015, 14, 177–194. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wang, H.; Zheng, X.; Xue, X.; Wang, B.; Wu, H.; Zhang, K.; Fan, S.; Wang, T.; Li, N. CpG/Poly (I: C) mixed adjuvant priming enhances the immunogenicity of a DNA vaccine against Eastern equine encephalitis virus in mice. Int. Immunopharmacol. 2014, 19, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P. Porcine deltacoronavirus infection and transmission in poultry, United States. Emerg Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riemenschneider, J.; Garrison, A.; Geisbert, J.; Jahrling, P.; Hevey, M.; Negley, D.; Schmaljohn, A.; Lee, J.; Hart, M.K.; Vanderzanden, L. Comparison of individual and combination DNA vaccines for B. anthracis, Ebola virus, Marburg virus and Venezuelan equine encephalitis virus. Vaccine 2003, 21, 4071–4080. [Google Scholar] [CrossRef] [Green Version]
- Perkins, S.D.; O’Brien, L.M.; Phillpotts, R.J. Boosting with an adenovirus-based vaccine improves protective efficacy against Venezuelan equine encephalitis virus following DNA vaccination. Vaccine 2006, 24, 3440–3445. [Google Scholar] [CrossRef]
- Dupuy, L.C.; Locher, C.P.; Paidhungat, M.; Richards, M.J.; Lind, C.M.; Bakken, R.; Parker, M.D.; Whalen, R.G.; Schmaljohn, C.S. Directed molecular evolution improves the immunogenicity and protective efficacy of a Venezuelan equine encephalitis virus DNA vaccine. Vaccine 2009, 27, 4152–4160. [Google Scholar] [CrossRef]
- Dupuy, L.C.; Richards, M.J.; Reed, D.S.; Schmaljohn, C.S. Immunogenicity and protective efficacy of a DNA vaccine against Venezuelan equine encephalitis virus aerosol challenge in nonhuman primates. Vaccine 2010, 28, 7345–7350. [Google Scholar] [CrossRef]
- Dupuy, L.C.; Richards, M.J.; Ellefsen, B.; Chau, L.; Luxembourg, A.; Hannaman, D.; Livingston, B.D.; Schmaljohn, C.S. A DNA vaccine for Venezuelan equine encephalitis virus delivered by intramuscular electroporation elicits high levels of neutralizing antibodies in multiple animal models and provides protective immunity to mice and nonhuman primates. Clin. Vaccine Immunol. 2011, 18, 707–716. [Google Scholar] [CrossRef] [Green Version]
- Suschak, J.J.; Bagley, K.; Six, C.; Shoemaker, C.J.; Kwilas, S.; Spik, K.W.; Dupuy, L.C.; Schmaljohn, C.S. The genetic adjuvant IL-12 enhances the protective efficacy of a DNA vaccine for Venezuelan equine encephalitis virus delivered by intramuscular injection in mice. Antivir. Res. 2018, 159, 113–121. [Google Scholar] [CrossRef]
- Hannaman, D.; Dupuy, L.C.; Ellefsen, B.; Schmaljohn, C.S. A Phase 1 clinical trial of a DNA vaccine for Venezuelan equine encephalitis delivered by intramuscular or intradermal electroporation. Vaccine 2016, 34, 3607–3612. [Google Scholar] [CrossRef]
- Tretyakova, I.; Lukashevich, I.S.; Glass, P.; Wang, E.; Weaver, S.; Pushko, P. Novel vaccine against Venezuelan equine encephalitis combines advantages of DNA immunization and a live attenuated vaccine. Vaccine 2013, 31, 1019–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bounds, C.E.; Terry, F.E.; Moise, L.; Hannaman, D.; Martin, W.D.; De Groot, A.S.; Suschak, J.J.; Dupuy, L.C.; Schmaljohn, C.S. An immunoinformatics-derived DNA vaccine encoding human class II T cell epitopes of Ebola virus, Sudan virus, and Venezuelan equine encephalitis virus is immunogenic in HLA transgenic mice. Hum. Vaccines Immunother. 2017, 13, 2824–2836. [Google Scholar] [CrossRef]
- Nagata, L.P.; Hu, W.-G.; Masri, S.A.; Rayner, G.A.; Schmaltz, F.L.; Das, D.; Wu, J.; Long, M.C.; Chan, C.; Proll, D. Efficacy of DNA vaccination against Western equine encephalitis virus infection. Vaccine 2005, 23, 2280–2283. [Google Scholar] [CrossRef] [PubMed]
- Gauci, P.J.; Wu, J.Q.; Rayner, G.A.; Barabé, N.D.; Nagata, L.P.; Proll, D.F. Identification of Western equine encephalitis virus structural proteins that confer protection after DNA vaccination. Clin. Vaccine Immunol. 2010, 17, 176–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dupuy, L.C.; Richards, M.J.; Livingston, B.D.; Hannaman, D.; Schmaljohn, C.S. A multiagent Alphavirus DNA vaccine delivered by intramuscular electroporation elicits robust and durable virus-specific immune responses in mice and rabbits and completely protects mice against lethal Venezuelan, Western, and Eastern equine encephalitis virus aerosol challenges. J. Immunol. Res. 2018, 2018, 8521060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundberg, L.; Carey, B.; Kehn-Hall, K. Venezuelan equine encephalitis virus capsid-the clever caper. Viruses 2017, 9, 279. [Google Scholar] [CrossRef] [Green Version]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Pascolo, S. Messenger RNA-based vaccines. Expert Opin. Biol. Ther. 2004, 4, 1285–1294. [Google Scholar] [CrossRef]
- Zhang, C.; Maruggi, G.; Shan, H.; Li, J. Advances in mRNA vaccines for infectious diseases. Front. Immunol. 2019, 10, 594. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Hogan, M.J.; Naradikian, M.S.; Parkhouse, K.; Cain, D.W.; Jones, L.; Moody, M.A.; Verkerke, H.P.; Myles, A.; Willis, E.; et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018, 215, 1571–1588. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Pelc, R.S.; Muramatsu, H.; Andersen, H.; DeMaso, C.R.; Dowd, K.A.; Sutherland, L.L.; Scearce, R.M.; Parks, R.; et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 2017, 543, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Weissman, D. Nucleoside modified mRNA vaccines for infectious diseases. Methods Mol. Biol. 2017, 1499, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsa, M.M.; Dupuy, L.C.; Beard, C.W.; Six, C.M.; Schmaljohn, C.S.; Mason, P.W.; Geall, A.J.; Ulmer, J.B.; Yu, D. Self-amplifying RNA vaccines for Venezuelan equine encephalitis virus induce robust protective immunogenicity in mice. Mol. Ther. 2019, 27, 850–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flower, D.R.; Macdonald, I.K.; Ramakrishnan, K.; Davies, M.N.; Doytchinova, I.A. Computer aided selection of candidate vaccine antigens. Immunome Res. 2010, 6 (Suppl. 2), S1. [Google Scholar] [CrossRef] [Green Version]
- Fischer, W.; Perkins, S.; Theiler, J.; Bhattacharya, T.; Yusim, K.; Funkhouser, R.; Kuiken, C.; Haynes, B.; Letvin, N.L.; Walker, B.D.; et al. Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 2007, 13, 100–106. [Google Scholar] [CrossRef]
- Korber, B.T.; Letvin, N.L.; Haynes, B.F. T-cell vaccine strategies for human immunodeficiency virus, the virus with a thousand faces. J. Virol. 2009, 83, 8300–8314. [Google Scholar] [CrossRef] [Green Version]
- Trolle, T.; McMurtrey, C.P.; Sidney, J.; Bardet, W.; Osborn, S.C.; Kaever, T.; Sette, A.; Hildebrand, W.H.; Nielsen, M.; Peters, B. The length distribution of class I-restricted T cell epitopes Is determined by both peptide supply and MHC allele-specific binding preference. J. Immunol. 2016, 196, 1480–1487. [Google Scholar] [CrossRef] [Green Version]
- Van Regenmortel, M.H. Virus species and virus identification: Past and current controversies. Infect. Genet. Evol. 2007, 7, 133–144. [Google Scholar] [CrossRef]
- Kong, W.P.; Wu, L.; Wallstrom, T.C.; Fischer, W.; Yang, Z.Y.; Ko, S.Y.; Letvin, N.L.; Haynes, B.F.; Hahn, B.H.; Korber, B.; et al. Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J. Virol. 2009, 83, 2201–2215. [Google Scholar] [CrossRef] [Green Version]
- Barouch, D.H.; O’Brien, K.L.; Simmons, N.L.; King, S.L.; Abbink, P.; Maxfield, L.F.; Sun, Y.H.; La Porte, A.; Riggs, A.M.; Lynch, D.M.; et al. Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 2010, 16, 319–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santra, S.; Liao, H.X.; Zhang, R.; Muldoon, M.; Watson, S.; Fischer, W.; Theiler, J.; Szinger, J.; Balachandran, H.; Buzby, A.; et al. Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat. Med. 2010, 16, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W. T cell-based strategies for HIV-1 vaccines. Hum. Vaccines Immunother. 2020, 16, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, K.E.; Wagh, K.; Korber, B.; Barouch, D.H. Vaccines and broadly neutralizing antibodies for HIV-1 prevention. Annu. Rev. Immunol. 2020, 38, 673–703. [Google Scholar] [CrossRef]
- Yusim, K.; Fischer, W.; Yoon, H.; Thurmond, J.; Fenimore, P.W.; Lauer, G.; Korber, B.; Kuiken, C. Genotype 1 and global hepatitis C T-cell vaccines designed to optimize coverage of genetic diversity. J. Gen. Virol. 2010, 91, 1194–1206. [Google Scholar] [CrossRef]
- Yusim, K.; Dilan, R.; Borducchi, E.; Stanley, K.; Giorgi, E.; Fischer, W.; Theiler, J.; Marcotrigiano, J.; Korber, B.; Barouch, D.H. Hepatitis C genotype 1 mosaic vaccines are immunogenic in mice and induce stronger T-cell responses than natural strains. Clin. Vaccine Immunol. 2013, 20, 302–305. [Google Scholar] [CrossRef] [Green Version]
- Fenimore, P.W.; Muhammad, M.A.; Fischer, W.M.; Foley, B.T.; Bakken, R.R.; Thurmond, J.R.; Yusim, K.; Yoon, H.; Parker, M.; Hart, M.K.; et al. Designing and testing broadly-protective filoviral vaccines optimized for cytotoxic T-lymphocyte epitope coverage. PLoS ONE 2012, 7, e44769. [Google Scholar] [CrossRef] [Green Version]
- Theiler, J.; Yoon, H.; Yusim, K.; Picker, L.J.; Fruh, K.; Korber, B. Epigraph: A vaccine design tool applied to an HIV therapeutic vaccine and a pan-Filovirus vaccine. Sci. Rep. 2016, 6, 33987. [Google Scholar] [CrossRef] [Green Version]
- Rahim, M.N.; Wee, E.G.; He, S.; Audet, J.; Tierney, K.; Moyo, N.; Hannoun, Z.; Crook, A.; Baines, A.; Korber, B.; et al. Complete protection of the BALB/c and C57BL/6J mice against Ebola and Marburg virus lethal challenges by pan-Filovirus T-cell epigraph vaccine. PLoS Pathog. 2019, 15, e1007564. [Google Scholar] [CrossRef]
- Abdul-Jawad, S.; Ondondo, B.; van Hateren, A.; Gardner, A.; Elliott, T.; Korber, B.; Hanke, T. Increased valency of conserved-mosaic vaccines enhances the breadth and depth of epitope recognition. Mol. Ther. 2016, 24, 375–384. [Google Scholar] [CrossRef] [Green Version]



| Target | Vaccine | Genes | Animal Model | Doses, Route | Induced Immune Responses | Challenge Route | % Survival Post-Challenge | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CI 1 | HI 2 | Homologous | Heterologous | |||||||
| Adenovirus | ||||||||||
| VEEV | RAd/VEEV#3 | E3-E2-6K | BALB/c mice | 3, IN 3 | NT 4 | Yes | Aerosol | 90 | 50–100 | [67] |
| 2, IN | NT | Yes | Aerosol | 10 | NT | [68] | ||||
| 3, IN | NT | Yes | Aerosol | 20 | NT | [69] | ||||
| RAd/VEEV#3 +CpG | E3-E2-6K | BALB/c mice | 2, IN | NT | Yes | Aerosol | 40 | NT | [68] | |
| RAd/VEEV#3 -CO | E3-E2-6K | BALB/c mice | 3, IN | NT | Yes | Aerosol | 90 | NT | [69] | |
| WEEV | Ad5-WEEV | E3-E2-6K-E1 | BALB/c mice | 2, IM 5 | NT | Yes | IN | 100 | NT | [70] |
| 1, IM | NT | Yes | IN | 100 | 88–100 | [71] | ||||
| Ad5-E1 | 6K-E1 | BALB/c mice | 1, IM | Yes | No | IN | 100 | 0–100 | [72] | |
| Eilat Virus | ||||||||||
| EEEV | EILV/EEEV | C-E3-E2-6K-E1 | CD-1 mice | 1, SC 6 | NT | Yes | IP 7 | 100 | NT | [73] |
| VEEV | EILV/VEEV | C-E3-E2-6K-E1 | CD-1 mice | 1, SC | NT | Yes | SC | NT | 100 | |
| EEEV, VEEV | EILV/EEEV, EILV/VEEV, EILV/CHIKV | C-E3-E2-6K-E1 and C-E2-E1 | CD-1 mice | 1, SC | NT | Yes | IP or SC | 80 | 90 | |
| Equine Herpes Virus | ||||||||||
| VEEV | rH_VEEV | E3-E2-6K-E1 | NIH Swiss mice | 2, SC | NT | Yes | SC | NT | 0–100 | [74] |
| Isfahan Virus | ||||||||||
| EEEV, VEEV | rISFV-VEEV/rISFV-EEEV | E3-E2-6K-E1 | CD-1 mice | 1, IM | NT | Yes | IP or SC | 100 | NT | [75] |
| Sindbis Virus | ||||||||||
| EEEV | SIN/NAEEEV | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | NT | Yes | IP | 80–100 | NT | [76] |
| Cynomolgus macaques | 1, SC | NT | Yes | Aerosol | 82 | NT | [77] | |||
| VEEV | SIN-83 | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | NT | Yes | SC | NT | 100 | [78] |
| 1, SC | Yes | Yes | IC 8 IN or SC | NT | 80–100 | [79] | ||||
| SAAR/TRD | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | Yes | Yes | IC, IN or SC | NT | 100 | ||
| Golden hamster | 1, SC | NT | NT | SC | NT | 100 | ||||
| SIN/TRD | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | Yes | Yes | IC, IN or SC | NT | 100 | ||
| Golden hamster | 1, SC | NT | NT | SC | NT | 100 | ||||
| SIN/ZPC | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | Yes | Yes | IC, IN or SC | 100 | NT | ||
| Golden hamster | 1, SC | NT | NT | SC | 100 | NT | ||||
| WEEV | SIN/CO92 | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | NT | Yes | IN | NT | 40–100 | [80] |
| SIN/SIN/McM or SIN/EEE/McM | C-E3-E2-6K-E1 | NIH Swiss mice | 1, SC | NT | Yes | IN | 100 | NT | ||
| Vaccinia Virus | ||||||||||
| EEEV | MVA-BN-E | E3-E2-6K-E1 | BALB/c mice | 2, SC | NT | Yes | IN | NT | 100 | [81] |
| VEEV | MVA-BN-V | E3-E2-6K-E1 | BALB/c mice | 2, SC | NT | Yes | IN | 100 | NT | |
| WEEV | MVA-BN-W | E3-E2-6K-E1 | BALB/c mice | 2, SC | NT | Yes | IN | NT | 100 | |
| EEEV, VEEV, WEEV | MVA-BN-W +E+V | E3-E2-6K-E1 | BALB/c mice | 2, SC | NT | Yes | IN | 90–100 | 60–100 | |
| Vesicular Stomatitis Virus | ||||||||||
| VEEV | rVSIV-VEEV | E3-E2-6K-E1 | CD-1 mice | 1, IM | NT | Yes | SC | 100 | NT | [75] |
| Target | Vaccine | Genes | Animal Model | Immunization Doses, Route | Induced Immune Responses | Challenge Route | % Survival Postchallenge | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CI 1 | HI 2 | Homologous | Heterologous | |||||||
| EEEV | pcDNA 3.1(+)-C-E | C-E3-E2-6K-E1 | BALB/c mice | 3×, IM 3 | Yes | Yes | NT 4 | NT | NT | [104] |
| VEEV | 26S | C-E3-E2-6K-E1 | BALB/c mice | 3×, gene gun | NT | Yes | Aerosol or SC 5 | 80–100 | NT | [105] |
| DNA-Ad | E3-E2-6K | BALB/c mice | 3×, gene gun and 1×, IN 6 | NT | Yes | Aerosol | 83 | NT | [106] | |
| AG2-5A7 | E3-E2-6K-E1 | BALB/c mice | 3×, ID 7 | NT | Yes | Aerosol | 80 | NT | [107] | |
| AG2-5A10 | E3-E2-6K-E1 | BALB/c mice | 3×, ID | NT | Yes | Aerosol | 70 | NT | ||
| AG4-1C7 | E3-E2-6K-E1 | BALB/c mice | 3×, ID | NT | Yes | Aerosol | 90 | NT | ||
| AG4-1G2 | E3-E2-6K-E1 | BALB/c mice | 3×, ID | NT | Yes | Aerosol | 100 | NT | ||
| VEEV DNA | C-E3-E2-6K-E1 | Cynomolgus macaques | 3×, gene gun | NT | Yes | Aerosol | 100 | NT | [108] | |
| VEEVCO | E3-E2-6K-E1 | BALB/c mice | 2×, IM-EP 8 | Yes | Yes | Aerosol | 100 | NT | [109] | |
| 2×, IM | Yes | Yes | Aerosol | 50–100 | NT | [110] | ||||
| Cynomolgus macaque | 2×, IM-EP | NT | Yes | Aerosol | 100 | NT | [109] | |||
| Human | 3×, IM-EP | NT | Yes | NT | NT | NT | [111] | |||
| VEEVCOCAP | C-E3-E2-6K-E1 | BALB/c mice | 2×, IM-EP | NT | Yes | Aerosol | 90 | NT | [109] | |
| pTC83 iDNA | Full-length cDNA | BALB/c mice | 1×, IM-EP | NT | Yes | SC | NT | 100 | [112] | |
| Multi-epitope DNA | Partial sequences of C-E2-E1 | BALB/c mice | 3×, IM-EP | Yes | Yes | NT | NT | NT | [113] | |
| HLA-DR3 mice | 2×, IM-EP | Yes | Yes | Aerosol | 20 | NT | ||||
| WEEV | pVXH-6 | C-E3-E2-6K-E1 | BALB/c mice | 4×, gene gun | Yes | No | IN | 100 | 50–62 | [114] |
| 3×, gene gun | NT | NT | IN | 100 | 50–63 | [115] | ||||
| pE3-E2-6K-E1 | E3-E2-6K-E1 | BALB/c mice | 3×, gene gun | NT | NT | IN | 100 | 88–100 | [115] | |
| pE3-E2 | E3-E2 | BALB/c mice | 3×, gene gun | NT | NT | IN | 0 | 0 | ||
| p6K-E1 | 6K-E1 | BALB/c mice | 3×, gene gun | NT | NT | IN | 100 | 0–75 | ||
| EEEV, VEEV, WEEV | 3-EEV | E3-E2-6K-E1 | BALB/c mice | 2×-3×, IM-EP | Yes | Yes | Aerosol | 100 | NT | [116] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stromberg, Z.R.; Fischer, W.; Bradfute, S.B.; Kubicek-Sutherland, J.Z.; Hraber, P. Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines 2020, 8, 273. https://doi.org/10.3390/vaccines8020273
Stromberg ZR, Fischer W, Bradfute SB, Kubicek-Sutherland JZ, Hraber P. Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines. 2020; 8(2):273. https://doi.org/10.3390/vaccines8020273
Chicago/Turabian StyleStromberg, Zachary R., Will Fischer, Steven B. Bradfute, Jessica Z. Kubicek-Sutherland, and Peter Hraber. 2020. "Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses" Vaccines 8, no. 2: 273. https://doi.org/10.3390/vaccines8020273
APA StyleStromberg, Z. R., Fischer, W., Bradfute, S. B., Kubicek-Sutherland, J. Z., & Hraber, P. (2020). Vaccine Advances against Venezuelan, Eastern, and Western Equine Encephalitis Viruses. Vaccines, 8(2), 273. https://doi.org/10.3390/vaccines8020273