Phosphorus Concentration in Water Affects the Biofilm Community and the Produced Amount of Extracellular Polymeric Substances in Reverse Osmosis Membrane Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Setup and Operational Parameters during Biofilm Growth
2.2. Forward Flushing and End of the Experiment
2.3. Optical Coherence Tomography
2.4. Biomass Quantification
2.4.1. Adenosine Triphosphate
2.4.2. Total Cell Count
2.4.3. Extraction and Quantification of Extracellular Polymeric Substances (EPS)
2.5. DNA Extraction and Illumina Sequencing
3. Results
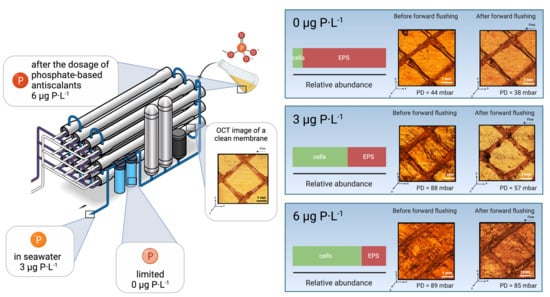
3.1. Feed Channel Pressure Drop Restoration and Visualization of Biofilm Removal
3.2. Biomass Characterization
3.3. Bacterial Diversity and Community Analysis
4. Discussion
4.1. Bacterial Adaptation to Phosphorus-Limiting Conditions
4.1.1. EPS Production
4.1.2. Phosphorus Accumulation, Regeneration, and Sequestration
4.1.3. Adhesin Protein Production
4.2. Homogenous Bacterial Community Composition throughout the Biofilm Height
4.3. Practical Implications and Future Research
5. Conclusions
- (i)
- (ii)
- For biofilms grown at lower phosphorus concentration conditions, the ratio of EPS production per bacterial cell was higher than for higher phosphorus concentrations (Figure 4).
- (iii)
- The relative abundance of main bacterial communities changed at varying dosed phosphorus concentrations in the feed water (Figure 6).
- (iv)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATP | Adenosine triphosphate |
| BSA | Bovine serum albumin |
| c-di-GMP | Cyclic dimeric guanosine monophosphate |
| EPS | Extracellular polymeric substances |
| MFS | Membrane fouling simulator |
| NCBI | National Center for Biotechnology Information |
| OCT | Optical Coherence Tomography |
| PAO | Polyphosphate-accumulating organisms |
| PBS | Phosphate-buffered saline solution |
| PCoA | Principal Coordinates Analysis |
| RDP | Ribosomal Database Project |
| RO | Reverse osmosis |
| SWRO | Seawater reverse osmosis |
| TCC | Total cell count |
References
- UNESCO. The United Nations World Water Development Report 2021: Valuing Water; UNESCO: Paris, France, 2021; p. 206. [Google Scholar]
- UN Environment. Global Environment Outlook—GEO-6: Healthy Planet, Healthy People; UN Environment: Nairobi, Kenya, 2019. [Google Scholar]
- Flemming, H.; Wingender, J. The biofilm matrix, nature reviews. Microbiology 2010, 8, 623–633. [Google Scholar]
- Vrouwenvelder, J.S.; Kruithof, J.C.; van Loosdrecht, M. Integrated approach for biofouling control. Water Sci. Technol. 2010, 62, 2477–2490. [Google Scholar] [CrossRef]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef] [PubMed]
- Burton, F.; Tchobanoglous, G.; Stensel, D. Wastewater Engineering: Treatment and Reuse, 4th ed.; Burton, F., Tchobanoglous, G., Stensel, H.D., Eds.; Tata McGraw-Hill Education Pvt. Ltd.: Delhi, India, 2010. [Google Scholar]
- Miettinen, I.T.; Vartiainen, T.; Martikainen, P.J. Phosphorus and bacterial growth in drinking water. Appl. Environ. Microbiol. 1997, 63, 3242–3245. [Google Scholar] [CrossRef] [Green Version]
- Chrzanowski, T.; Kyle, M. Ratios of carbon, nitrogen and phosphorus in Pseudomonas fluorescens as a model for bacterial element ratios and nutrient regeneration. Aquat. Microb. Ecol. 1996, 10, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Javier, L.; Farhat, N.M.; Desmond, P.; Linares, R.V.; Bucs, S.; Kruithof, J.C.; Vrouwenvelder, J.S. Biofouling control by phospho-rus limitation strongly depends on the assimilable organic carbon concentration. Water Res. 2020, 183, 116051. [Google Scholar] [CrossRef] [PubMed]
- Farhat, N.M.; Javier, L.; van Loosdrecht, M.C.M.; Kruithof, J.C.; Vrouwenvelder, J.S. Role of feed water biodegradable substrate concentration on biofouling: Biofilm characteristics, membrane performance and cleanability. Water Res. 2019, 150, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, J.D.; Kennedy, M.D.; Amy, G.; Schippers, J.C. Phosphate limitation in reverse osmosis: An option to control biofouling? Desalin. Water Treat. 2009, 5, 198–206. [Google Scholar] [CrossRef] [Green Version]
- Vrouwenvelder, J.S.; Beyer, F.; Dahmani, K.; Hasan, N.; Galjaard, G.; Kruithof, J.C.; van Loosdrecht, M.C.M. Phosphate limitation to control biofouling. Water Res. 2010, 44, 3454–3466. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-M.; Kim, S.-J.; Kim, L.H.; Shin, M.S.; Yu, H.-W.; Kim, I.S. Effects of phosphate limitation in feed water on biofouling in forward osmosis (FO) process. Desalination 2014, 349, 51–59. [Google Scholar] [CrossRef]
- Danhorn, T.; Hentzer, M.; Givskov, M.; Parsek, M.R.; Fuqua, C. Phosphorus Limitation Enhances Biofilm Formation of the Plant Pathogen Agrobacterium tumefaciens through the PhoR-PhoB Regulatory System. J. Bacteriol. 2004, 186, 4492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmond, P.; Best, J.P.; Morgenroth, E.; Derlon, N. Linking composition of extracellular polymeric substances (EPS) to the physical structure and hydraulic resistance of membrane bio films. Water Res. 2018, 132, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullan, A.; Quinn, J.P.; McGrath, J.W. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low-pH conditions. Microb. Ecol. 2002, 44, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Song, O.-R.; Lee, S.-J.; Lee, Y.-S.; Lee, S.-C.; Kim, K.-K.; Choi, Y.-L. Solubilization of insoluble inorganic phosphate by Burkholderia cepacia DA23 isolated from cultivated soil. Braz. J. Microbiol. 2008, 39, 151. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Q.; Wu, X.; Wang, J.; Ding, X. Phosphate Solubilization and Gene Expression of Phosphate-Solubilizing Bacterium Burkholderia multivorans WS-FJ9 under Different Levels of Soluble Phosphate. J. Microbiol. Biotechnol. 2017, 27, 844–855. [Google Scholar] [CrossRef] [PubMed]
- JPoindexter, L.S.; Cohen-Bazire, G. The fine structure of stalked bacteria belonging to the family Caulobacteriaceae. J. Cell Biol. 1964, 23, 587. [Google Scholar] [CrossRef]
- Poindexter, J.S. The role of calcium in stalk development and in phosphate acquisition in Caulobacter crescentus. Arch. Microbiol. 1984, 138, 140–152. [Google Scholar] [CrossRef]
- Schmidt, J.M.; Stanier, R.Y. The development of cellular stalks in bacteria. J. Cell Biol. 1966, 28, 423–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonin, M.; Quardokus, E.M.; O’Donnol, D.; Maddock, J.; Brun, Y.V. Regulation of Stalk Elongation by Phosphate in Caulobacter crescentus. J. Bacteriol. 2000, 182, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, N.; Suppiger, A.; Steiner, E.; Pessi, G.; Kaever, V.; Fazli, M.M.; Tolker-Nielsen, T.; Jenal, U.; Eberl, L. High intracellular c-di-GMP levels antagonize quorum sensing and virulence gene expression in Burkholderia cenocepacia H111. Microbiology 2017, 163, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Newell, P.D.; Boyd, C.D.; Sondermann, H.; O’Toole, G.A. A c-di-GMP Effector System Controls Cell Adhesion by Inside-Out Signaling and Surface Protein Cleavage. PLoS Biol. 2011, 9, e1000587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navarro, M.; Newell, P.D.; Krasteva, P.V.; Chatterjee, D.; Madden, D.R.; O’Toole, G.A.; Sondermann, H. Structural Basis for c-di-GMP-Mediated Inside-Out Signaling Controlling Periplasmic Proteolysis. PLoS Biol. 2011, 9, e1000588. [Google Scholar] [CrossRef] [PubMed]
- Monds, R.D.; Newell, P.D.; Gross, R.H.; O’Toole, G.A. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudo-monas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007, 63, 656–679. [Google Scholar] [CrossRef] [PubMed]
- Kalia, D.; Merey, G.; Nakayama, S.; Zheng, Y.; Zhou, J.; Luo, Y.; Guo, M.; Roembke, B.T.; Sintim, H.O. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chemi. Soc. Rev. 2013, 42, 305–341. [Google Scholar] [CrossRef]
- Tejedor-Sanz, S.; Fernández-Labrador, P.; Hart, S.; Torres, C.I.; Esteve-Núñez, A. Geobacter dominates the inner layers of a stratified biofilm on a fluidized anode during brewery wastewater treatment. Front. Microbiol. 2018, 9, 378. [Google Scholar] [CrossRef]
- Almstrand, R.; Persson, F.; Daims, H.; Ekenberg, M.; Christensson, M.; Wilén, B.-M.; Sörensson, F.; Hermansson, M. Three-Dimensional Stratification of Bacterial Biofilm Populations in a Moving Bed Biofilm Reactor for Nitritation-Anammox. Int. J. Mol. Sci. 2014, 15, 2191–2206. [Google Scholar] [CrossRef] [Green Version]
- Javier, L.; Farhat, N.M.; Vrouwenvelder, J.S. Enhanced hydraulic cleanability of biofilms developed under a low phosphorus concentration in reverse osmosis membrane systems. Water Res. X 2020, 10, 100085. [Google Scholar] [CrossRef]
- Vrouwenvelder, J.S.; Hinrichs, C.; Van Der Meer, W.G.; van Loosdrecht, M.; Kruithof, J.C. Pressure drop increase by biofilm accumulation in spiral wound RO and NF membrane systems: Role of substrate concentration, flow velocity, substrate load and flow direction. Biofouling 2009, 25, 543–555. [Google Scholar] [CrossRef]
- Abushaban, A.; Salinas-Rodriguez, S.G.; Kapala, M.; Pastorelli, D.; Schippers, J.C.; Mondal, S.; Goueli, S.; Kennedy, M.D. Moni-toring Biofouling Potential Using ATP-Based Bacterial Growth Potential in SWRO Pre-Treatment of a Full-Scale Plant. Membranes 2020, 10, 360. [Google Scholar] [CrossRef]
- Sanawar, H.; Siddiqui, A.; Bucs, S.; Farhat, N.; van Loosdrecht, M.; Kruithof, J.; Vrouwenvelder, J. Applicability of short-term accelerated biofouling studies to predict long-term biofouling accumulation in reverse osmosis membrane systems. Desalin. Water Treat. 2017, 97, 72–78. [Google Scholar] [CrossRef]
- Farhat, N.; Kim, L.H.; Vrouwenvelder, J.S. Online characterization of bacterial processes in drinking water systems. npj Clean Water 2020, 3, 16. [Google Scholar] [CrossRef] [Green Version]
- Pearson, J.L.; Michael, P.R.; Ghaffour, N.; Missimer, T.M. Economics and Energy Consumption of Brackish Water Reverse Osmosis Desalination: Innovations and Impacts of Feedwater Quality. Membranes 2021, 11, 616. [Google Scholar] [CrossRef]
- Vrouwenvelder, J.S.; Bakker, S.M.; Wessels, L.P.; van Paassen, J.A.M. The Membrane Fouling Simulator as a new tool for bio-fouling control of spiral-wound membranes. Desalination 2007, 204, 170–174. [Google Scholar] [CrossRef]
- Siebdrath, N.; Farhat, N.; Ding, W.; Kruithof, J.; Vrouwenvelder, J.S. Impact of membrane biofouling in the sequential development of performance indicators: Feed channel pressure drop, permeability, and salt rejection. J. Membr. Sci. 2019, 585, 199–207. [Google Scholar] [CrossRef]
- Vrouwenvelder, J.S.; Bakker, S.M.; Cauchard, M.; le Grand, R.; Apacandié, M.; Idrissi, M.; Lagrave, S.; Wessels, L.P.; van Paasen, J.A.M.; Kruithof, J.C.; et al. The membrane fouling simulator: A suitable tool for prediction and charactersation of membrane fouling. Water Sci. Technol. 2007, 55, 197–205. [Google Scholar] [CrossRef]
- Bucs, S.S.; Farhat, N.; Siddiqui, A.; Linares, R.V.; Radu, A.; Kruithof, J.C.; Vrouwenvelder, J.S. Development of a setup to enable stable and accurate flow conditions for membrane biofouling studies. Desalin. Water Treat. 2015, 57, 12893–12901. [Google Scholar] [CrossRef]
- Dreszer, C.; Flemming, H.-C.; Zwijnenburg, A.; Kruithof, J.C.; Vrouwenvelder, J.S. Impact of biofilm accumulation on trans-membrane and feed channel pressure drop: Effects of crossflow velocity, feed spacer and biodegradable nutrient. Water Res. 2014, 50, 200–211. [Google Scholar] [CrossRef]
- Abushaban, A.; Salinas-Rodriguez, S.G.; Dhakal, N.; Schippers, J.C.; Kennedy, M.D. Assessing pretreatment and seawater re-verse osmosis performance using an ATP-based bacterial growth potential method. Desalination 2019, 467, 210–218. [Google Scholar] [CrossRef]
- Kuhn, R.; Vornholt, C.; Preuß, V.; Bryant, I.M.; Martienssen, M. Aminophosphonates in Nanofiltration and Reverse Osmosis Permeates. Membranes 2021, 11, 446. [Google Scholar] [CrossRef] [PubMed]
- Sevcenco, A.-M.; Paravidino, M.; Vrouwenvelder, H.; Wolterbeek, H.T.; Van Loosdrecht, M.C.; Hagen, W.R. Phosphate and arsenate removal efficiency by thermostable ferritin enzyme from Pyrococcus furiosus using radioisotopes. Water Res. 2015, 76, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neu, L.; Proctor, C.R.; Walser, J.-C.; Hammes, F. Small-Scale Heterogeneity in Drinking Water Biofilms. Front. Microbiol. 2019, 10, 2446. [Google Scholar] [CrossRef] [PubMed]
- Farhat, N.; Hammes, F.; Prest, E.; Vrouwenvelder, J. A uniform bacterial growth potential assay for different water types. Water Res. 2018, 142, 227–235. [Google Scholar] [CrossRef]
- Dhakal, N.; Salinas-Rodriguez, S.G.; Ampah, J.; Schippers, J.C.; Kennedy, M.D. Measuring Biofouling Potential in SWRO Plants with a Flow-Cytometry-Based Bacterial Growth Potential Method. Membranes 2021, 11, 76. [Google Scholar] [CrossRef] [PubMed]
- Hammes, F.A.; Egli, T. New Method for Assimilable Organic Carbon Determination Using Flow-Cytometric Enumeration and a Natural Microbial Consortium as Inoculum. Environ. Sci. Technol. 2005, 39, 3289–3294. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Fang, H.H. Extraction of extracellular polymeric substances (EPS) of sludges. J. Biotechnol. 2002, 95, 249–256. [Google Scholar] [CrossRef]
- Masuko, T.; Minami, A.; Iwasaki, N.; Majima, T.; Nishimura, S.-I.; Lee, Y.C. Carbohydrate analysis by a phenol–sulfuric acid method in microplate format. Anal. Biochem. 2005, 339, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Loferer-Krößbacher, M.; Klima, J.; Psenner, R. Determination of Bacterial Cell Dry Mass by Transmission Electron Microscopy and Densitometric Image Analysis. Appl. Environ. Microbiol. 1998, 64, 688–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases de-tection of SAR11 bacterioplankton. Aquat. Micro. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Illumina. 16S Metagenomic Sequencing Library Preparation; Illumina: San Diego, CA, USA, 2015. [Google Scholar]
- Wang, Q.; Garrity, G.; Tiedje, J.; Cole, J. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Legendre, P.; Gallagher, E.D. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef]
- Lünsmann, V.; Kappelmeyer, U.; Benndorf, R.; Martinez-Lavanchy, P.M.; Taubert, A.; Adrian, L.; Duarte, M.; Pieper, D.H.; von Bergen, M.; Müller, J.A.; et al. In situprotein-SIP highlights Burkholderiaceae as key players degrading toluene by para ring hydroxylation in a constructed wetland model. Environ. Microbiol. 2016, 18, 1176–1186. [Google Scholar] [CrossRef] [PubMed]
- Szabó, E.; Liébana, R.; Hermansson, M.; Modin, O.; Persson, F.; Wilén, B.-M. Microbial Population Dynamics and Ecosystem Functions of Anoxic/Aerobic Granular Sludge in Sequencing Batch Reactors Operated at Different Organic Loading Rates. Front. Microbiol. 2017, 8, 770. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.W.; Ning, T.T.; Chen, Y.P.; Cheng, X.X.; Ren, N.Q. Bioaugmentation of Aerobic Granular Sludge with the Addition of a Bioflocculant-Producing Consortium. Adv. Mater. Res. 2013, 726–731, 2530–2535. [Google Scholar] [CrossRef]
- Weissbrodt, D.G.; Shani, N.; Holliger, C. Linking bacterial population dynamics and nutrient removal in the granular sludge biofilm ecosystem engineered for wastewater treatment. FEMS Microbiol. Ecol. 2014, 88, 579–595. [Google Scholar] [CrossRef] [Green Version]
- Jinhua, P.; Fukushi, K.; Yamamoto, K. Bacterial Community Structure on Membrane Surface and Characteristics of Strains Isolated from Membrane Surface in Submerged Membrane Bioreactor. Sep. Sci. Technol. 2006, 41, 1527–1549. [Google Scholar] [CrossRef]
- Jones, R.A.; Shropshire, H.; Zhao, C.; Murphy, A.; Lidbury, I.; Wei, T.; Scanlan, D.J.; Chen, Y. Phosphorus stress induces the synthesis of novel glycolipids in Pseudomonas aeruginosa that confer protection against a last-resort antibiotic. ISME J. 2021, 15, 3303–3314. [Google Scholar] [CrossRef] [PubMed]
- Bains, M.; Fernández, L.; Hancock, R.E.W. Phosphate Starvation Promotes Swarming Motility and Cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 6762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaborin, A.; Romanowski, K.; Gerdes, S.; Holbrook, C.; Lepine, F.; Long, J.; Poroyko, V.; Diggle, S.P.; Wilke, A.; Righetti, K.; et al. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Nat. Acad. Sci. USA 2009, 106, 6327–6332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jafari, M.; Derlon, N.; Desmond, P.; van Loosdrecht, M.C.M.; Morgenroth, E.; Picioreanu, C. Biofilm compressibility in ultrafiltration: A relation between biofilm morphology, mechanics and hydraulic resistance. Water Res. 2019, 157, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hu, S.; Sharma, K.R.; Ni, B.-J.; Yuan, Z. Stratified Microbial Structure and Activity in Sulfide- and Methane-Producing Anaerobic Sewer Biofilms. Appl. Environ. Microbiol. 2014, 80, 7042–7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Liu, J.; Bai, R. Membrane aerated biofilm reactors: A brief current review. Recent Patents Biotechnol. 2008, 2, 88–93. [Google Scholar] [CrossRef]
- Li, T.; Liu, J. Factors affecting performance and functional stratification of membrane-aerated biofilms with a counter-diffusion configuration. RSC Adv. 2019, 9, 29337–29346. [Google Scholar] [CrossRef] [Green Version]
- Ricardo, A.R.; Carvalho, G.; Velizarov, S.; Crespo, J.; Reis, M.A. Kinetics of nitrate and perchlorate removal and biofilm stratification in an ion exchange membrane bioreactor. Water Res. 2012, 46, 4556–4568. [Google Scholar] [CrossRef] [PubMed]
- Desmond, P.; Böni, L.; Fischer, P.; Morgenroth, E.; Derlon, N. Stratification in the physical structure and cohesion of membrane biofilms—Implications for hydraulic resistance. J. Membr. Sci. 2018, 564, 897–904. [Google Scholar] [CrossRef]
- Ghaffour, N.; Missimer, T.M.; Amy, G.L. Technical review and evaluation of the economics of water desalination: Current and future challenges for better water supply sustainability. Desalination 2013, 309, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Jafari, M.; Vanoppen, M.; van Agtmaal, J.; Cornelissen, E.; Vrouwenvelder, J.; Verliefde, A.; van Loosdrecht, M.; Picioreanu, C. Cost of fouling in full-scale reverse osmosis and nanofiltration installations in the Netherlands. Desalination 2020, 500, 114865. [Google Scholar] [CrossRef]
- Araújo, P.; Miller, D.; Correia, P.; van Loosdrecht, M.; Kruithof, J.; Freeman, B.; Paul, D.; Vrouwenvelder, J. Impact of feed spacer and membrane modification by hydrophilic, bactericidal and biocidal coating on biofouling control. Desalination 2012, 295, 1–10. [Google Scholar] [CrossRef]
- Sanawar, H.; Pinel, I.; Farhat, N.; Bucs, S.; Zlopasa, J.; Kruithof, J.; Witkamp, G.-J.; van Loosdrecht, M.; Vrouwenvelder, J. Enhanced biofilm solubilization by urea in reverse osmosis membrane systems. Water Res. X 2018, 1, 100004. [Google Scholar] [CrossRef]
- Nava-Ocampo, M.F.; Al Fuhaid, L.; Verpoorte, R.; Choi, Y.H.; van Loosdrecht, M.C.; Vrouwenvelder, J.S.; Witkamp, G.J.; Farinha, A.S.; Bucs, S.S. Natural deep eutectic solvents as biofilm structural breakers. Water Res. 2021, 201, 117323. [Google Scholar] [CrossRef] [PubMed]
- Andes, K.; Bartels, C.R.; Liu, E.; Sheehy, N. Methods for Enhanced Cleaning of Fouled RO Elements. 2013. Available online: http://membranes.com/wp-content/uploads/Documents/Technical-Papers/Application/Ind/METHODS-FOR-ENHANCED-CLEANING-OF-FOULED-RO-ELEMENTS.pdf (accessed on 25 September 2019).
- Alnajjar, H.; Tabatabai, A.; Alpatova, A.; Leiknes, T.; Ghaffour, N. Organic fouling control in reverse osmosis (RO) by effective membrane cleaning using saturated CO2 solution. Sep. Purif. Technol. 2021, 264, 118410. [Google Scholar] [CrossRef]
- Wibisono, Y.; Ahmad, F.; Cornelissen, E.; Kemperman, A.J.; Nijmeijer, D.C. Dominant factors controlling the efficiency of two-phase flow cleaning in spiral-wound membrane elements. Desalin. Water Treat. 2015, 57, 17625–17636. [Google Scholar] [CrossRef] [Green Version]
- Vrouwenvelder, J.; Buiter, J.; Riviere, M.; Van Der Meer, W.; Van Loosdrecht, M.; Kruithof, J. Impact of flow regime on pressure drop increase and biomass accumulation and morphology in membrane systems. Water Res. 2010, 44, 689–702. [Google Scholar] [CrossRef]
- Leon, F.; Ramos, A.; Perez-Baez, S.O. Optimization of Energy Efficiency, Operation Costs, Carbon Footprint and Ecological Footprint with Reverse Osmosis Membranes in Seawater Desalination Plants. Membranes 2021, 11, 781. [Google Scholar] [CrossRef] [PubMed]







| Dosed Carbon Concentration (µg C·L−1) as Glucose | Dosed Nitrogen Concentration (µg N·L−1) as Sodium Nitrate | Dosed Phosphorus Concentration (µg P·L−1) as Sodium Phosphate | C:N:P Ratio | Forward Flushing |
|---|---|---|---|---|
| 125 | 25 | 0 | 100:20:0.24 * | No |
| 0 | 100:20:0.24 * | Yes | ||
| 3 | 100:20:2.4 * | No | ||
| 3 | 100:20:2.4 * | Yes | ||
| 6 | 100:20:4.8 * | No | ||
| 6 | 100:20:4.8 * | Yes |
| Study | Dosed Phosphorus Concentration | 6 µg P·L−1 | 3 µg P·L−1 | 0 µg P·L−1 |
|---|---|---|---|---|
| Adenosine triphosphate (ng·cm−2) | Before forward flushing | 88.52 ± 6.55 | 24.54 ± 5.04 | 2.44 ± 0.41 |
| After forward flushing | 61.07 ± 2.75 | 4.48 ± 0.49 | 1.34 ± 0.45 | |
| Total cell count (×107 cells cm−2) | Before forward flushing | 8.01 ± 0.28 | 2.96 ± 0.17 | 0.11 ± 0.01 |
| After forward flushing | 5.70 ± 0.20 | 0.97 ± 0.02 | 0.06 ± 0.00 | |
| Proteins (µg·cm−2) | Before forward flushing | 8.22 ± 1.05 | 7.21 ± 0.58 | 3.54 ± 0.12 |
| After forward flushing | 7.77 ± 0.89 | 5.39 ± 1.16 | 3.50 ± 0.03 | |
| Extracellular polymeric substances in terms of proteins and carbohydrates (µg·cm−2) | Before forward flushing | 17.22 ± 3.05 | 12.21 ± 1.58 | 8.39 ± 0.37 |
| After forward flushing | 13.77 ± 1.89 | 9.39 ± 2.16 | 7.72 ± 1.40 |
| This Experiment | Literature | ||
|---|---|---|---|
| Dosed Phosphorus Concentration | Bacterial Family and Class Ordered from Higher to Lower Percentage of Relative Abundance | Effect on Biofilm Development Under Phosphate-Limiting Conditions | Reference |
| 6 µg P·L−1 | Burkholderiaceae (betaprotobacteria) | Adhesin protein production and P accumulation | [17,24,57] |
| Pseudomonadaceae (gammaprotobacteria) | Increase in quorum-sensing signals that promote biofilm formation, P regeneration | [8,62,63,64] | |
| Sphingomonadaceae (alphaproteobacteria) | EPS production | [58,59,60,61] | |
| 3 µg P·L−1 | Sphingomonadaceae (alphaproteobacteria) | EPS production | [58,59,60,61] |
| Burkholderiaceae (betaprotobacteria) | Adhesin protein production and P accumulation | [17,24,57] | |
| 0 µg P·L−1 | Sphingomonadaceae (alphaproteobacteria) | EPS production | [58,59,60,61] |
| Burkholderiaceae (betaprotobacteria) | Adhesin protein production and P accumulation | [17,24,57] | |
| Caulobacteraceae (alphaproteobacteria) | Phosphorus sequestration | [20,21,22,23] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Javier, L.; Pulido-Beltran, L.; Kruithof, J.; Vrouwenvelder, J.S.; Farhat, N.M. Phosphorus Concentration in Water Affects the Biofilm Community and the Produced Amount of Extracellular Polymeric Substances in Reverse Osmosis Membrane Systems. Membranes 2021, 11, 928. https://doi.org/10.3390/membranes11120928
Javier L, Pulido-Beltran L, Kruithof J, Vrouwenvelder JS, Farhat NM. Phosphorus Concentration in Water Affects the Biofilm Community and the Produced Amount of Extracellular Polymeric Substances in Reverse Osmosis Membrane Systems. Membranes. 2021; 11(12):928. https://doi.org/10.3390/membranes11120928
Chicago/Turabian StyleJavier, Luisa, Laura Pulido-Beltran, Joop Kruithof, Johannes S. Vrouwenvelder, and Nadia M. Farhat. 2021. "Phosphorus Concentration in Water Affects the Biofilm Community and the Produced Amount of Extracellular Polymeric Substances in Reverse Osmosis Membrane Systems" Membranes 11, no. 12: 928. https://doi.org/10.3390/membranes11120928
APA StyleJavier, L., Pulido-Beltran, L., Kruithof, J., Vrouwenvelder, J. S., & Farhat, N. M. (2021). Phosphorus Concentration in Water Affects the Biofilm Community and the Produced Amount of Extracellular Polymeric Substances in Reverse Osmosis Membrane Systems. Membranes, 11(12), 928. https://doi.org/10.3390/membranes11120928