Reagent-Free Immobilization of Industrial Lipases to Develop Lipolytic Membranes with Self-Cleaning Surfaces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Enzyme Immobilization
2.3. Enzyme Activity
2.4. RSM Design
2.5. Fouling and Self-Cleaning
2.6. Characterization
3. Results and Discussion
3.1. Enzyme Immobilization

3.2. Characterization
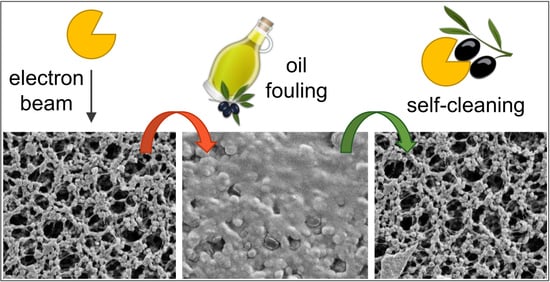
3.3. Fouling and Self-Cleaning
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arbige, M.V.; Shetty, J.K.; Chotani, G.K. Industrial Enzymology: The Next Chapter. Trends Biotechnol. 2019, 37, 1355–1366. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Christena, L.R.; Rajaram, Y.R. Enzyme immobilization: An overview on techniques and support materials. 3 Biotech 2013, 3, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldon, R.A.; Pelt, S.V. Enzyme immobilisation in biocatalysis: Why, what and how? Chem. Soc.Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prazeres, D.M.F.; Cabral, J.M.S. Enzymatic membrane bioreactors and their applications. Enzym. Microb. Technol. 1994, 16, 738–750. [Google Scholar] [CrossRef]
- Giorno, L.; Drioli, E. Biocatalytic membrane reactors: Applications and perspectives. Trends Biotechnol. 2000, 18, 339–349. [Google Scholar] [CrossRef]
- Jochems, P.; Satyawali, Y.; Diels, L.; Dejonghe, W. Enzyme immobilization on/in polymeric membranes: Status, challenges and perspectives in biocatalytic membrane reactors (BMRs). Green Chem. 2011, 13, 1609. [Google Scholar] [CrossRef]
- Sitanggang, A.B.; Drews, A.; Kraume, M. Enzymatic membrane reactors: Designs, applications, limitations and outlook. Chem. Eng. Processing Process Intensif. 2021, 168, 108729. [Google Scholar] [CrossRef]
- Dong, J.; Ning, W.; Liu, W.; Bruening, M.L. Limited proteolysis in porous membrane reactors containing immobilized trypsin. Analyst 2017, 142, 2578–2586. [Google Scholar] [CrossRef]
- Gupta, S. Comparative study on hydrolysis of oils by lipase immobilized biocatalytic PS membranes using biphasic enzyme membrane reactor. J. Environ. Chem. Eng. 2016, 4, 1797–1809. [Google Scholar] [CrossRef]
- Chhabra, M.; Mishra, S.; Sreekrishnan, T.R. Laccase/mediator assisted degradation of triarylmethane dyes in a continuous membrane reactor. J. Biotechnol. 2009, 143, 69–78. [Google Scholar] [CrossRef]
- Cazes, M.D.; Belleville, M.P.; Petit, E.; Salomo, M.; Bayer, S.; Czaja, R.; Gunzburg, J.D.; Sanchez-Marcano, J. Erythromycin degradation by esterase (EreB) in enzymatic membrane reactors. Biochem. Eng. J. 2016, 114, 70–78. [Google Scholar] [CrossRef]
- Algieri, C.; Donato, L.; Bonacci, P.; Giorno, L. Tyrosinase immobilised on polyamide tubular membrane for the L-DOPA production: Total recycle and continuous reactor study. Biochem. Eng. J. 2012, 66, 14–19. [Google Scholar] [CrossRef]
- Giorno, L.; Drioli, E.; Carvoli, G.; Cassano, A.; Donato, L. Study of an enzyme membrane reactor with immobilized fumarase for production of L-malic acid. Biotechnol. Bioeng. 2001, 72, 77–84. [Google Scholar] [CrossRef]
- Barbhuiya, N.H.; Misra, U.; Singh, S.P. Biocatalytic membranes for combating the challenges of membrane fouling and micropollutants in water purification: A review. Chemosphere 2022, 286, 131757. [Google Scholar] [CrossRef]
- Pekgenc, E.; Gul, B.Y.; Vatanpour, V.; Koyuncu, I. Biocatalytic membranes in anti-fouling and emerging pollutant degradation applications: Current state and perspectives. Sep. Purif. Technol. 2022, 282, 120098. [Google Scholar] [CrossRef]
- Schulze, A.; Breite, D.; Kim, Y.; Schmidt, M.; Thomas, I.; Went, M.; Fischer, K.; Prager, A. Bio-Inspired Polymer Membrane Surface Cleaning. Polymers 2017, 9, 97. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Breite, D.; Thomas, I.; Went, M.; Prager, A.; Schulze, A. Polymer membranes for active degradation of complex fouling mixtures. J. Membr. Sci. 2018, 563, 481–491. [Google Scholar] [CrossRef]
- Es, I.; Vieira, J.D.; Amaral, A.C. Principles, techniques, and applications of biocatalyst immobilization for industrial application. Appl. Microbiol. Biotechnol. 2015, 99, 2065–2082. [Google Scholar] [CrossRef]
- Schulze, A.; Marquardt, B.; Kaczmarek, S.; Schubert, R.; Prager, A.; Buchmeiser, M.R. Electron Beam-Based Functionalization of Poly(ethersulfone) Membranes. Macromol. Rapid Commun. 2010, 31, 467–472. [Google Scholar] [CrossRef]
- Schulze, A.; Marquardt, B.; Went, M.; Prager, A.; Buchmeiser, M.R. Electron beam-based functionalization of polymer membranes. Water Sci. Technol. 2012, 65, 574–580. [Google Scholar] [CrossRef]
- Becker-Jahn, J.; Griebel, J.; Glaß, S.; Langowski, P.; Nieß, S.; Schulze, A. Photoactive polymer membranes for degradation of pharmaceuticals from water. Catal. Today 2021, 364, 256–262. [Google Scholar] [CrossRef]
- Schulze, A.; Maitz, M.F.; Zimmermann, R.; Marquardt, B.; Fischer, M.; Werner, C.; Went, M.; Thomas, I. Permanent surface modification by electron-beam-induced grafting of hydrophilic polymers to PVDF membranes. RSC Adv. 2013, 3, 22518–22526. [Google Scholar] [CrossRef]
- Reinhardt, A.; Thomas, I.; Schmauck, J.; Giernoth, R.; Schulze, A.; Neundorf, I. Electron Beam Immobilization of Novel Antimicrobial, Short Peptide Motifs Leads to Membrane Surfaces with Promising Antibacterial Properties. J. Funct. Biomater. 2018, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Latif, A.A.; Prager, A.; Glaser, R.; Schulze, A. Highly Efficient One-Step Protein Immobilization on Polymer Membranes Supported by Response Surface Methodology. Front. Chem. 2021, 9, 804698. [Google Scholar] [CrossRef]
- Starke, S.; Went, M.; Prager, A.; Schulze, A. A novel electron beam-based method for the immobilization of trypsin on poly(ethersulfone) and poly(vinylidene fluoride) membranes. React. Funct. Polym. 2013, 73, 698–702. [Google Scholar] [CrossRef]
- Jahangiri, E.; Reichelt, S.; Thomas, I.; Hausmann, K.; Schlosser, D.; Schulze, A. Electron beam-induced immobilization of laccase on porous supports for waste water treatment applications. Molecules 2014, 19, 11860–11882. [Google Scholar] [CrossRef] [Green Version]
- Kuzmič, P. DynaFit—A Software Package for Enzymology. Methods Enzymol. 2009, 467, 247–280. [Google Scholar]
- Kuzmic, P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Anal. Biochem. 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Gonzalez, M.L.R.; Cornell-Kennon, S.; Schaefer, E.; Kuzmic, P. An algebraic model to determine substrate kinetic parameters by global nonlinear fit of progress curves. Anal. Biochem. 2017, 518, 16–24. [Google Scholar] [CrossRef]
- Golicnik, M. The integrated Michaelis-Menten rate equation: Deja vu or vu jade? J. Enzym. Inhib. Med. Chem. 2013, 28, 879–893. [Google Scholar] [CrossRef] [PubMed]
- Schnell, S.; Mendoza, C. Closed Form Solution for Time-dependent Enzyme Kinetics. J. Theor. Biol. 1997, 187, 207–212. [Google Scholar] [CrossRef]
- Box, G.E.P.; Wilson, K.B. On the Experimental Attainment of Optimum Conditions. In Breakthroughs in Statistics; Kotz, S., Johnson, N.L., Eds.; Springer Series in Statistics; Springer: New York, NY, USA, 1951; Volume 13, pp. 270–310. [Google Scholar] [CrossRef]
- Derringer, G.; Suich, R. Simultaneous Optimization of Several Response Variables. J. Qual. Technol. 2018, 12, 214–219. [Google Scholar] [CrossRef]
- Schulze, A.; Stoelzer, A.; Striegler, K.; Starke, S.; Prager, A. Biocatalytic Self-Cleaning Polymer Membranes. Polymers 2015, 7, 1837–1849. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Anderson, M.J.; Whitcomb, P.J. Design of Experiments. In Kirk-Othmer Encyclopedia of Chemical Technology; John Wiley & Sons: Hoboken, NJ, USA, 2010. [Google Scholar]
- Maurich, V.; Zacchigna, M.; Pitotti, A. P-nitrophenyllaurate: A substrate for the high-performance liquid chromatographic determination of lipase activity. J. Chromatogr. 1991, 566, 453–459. [Google Scholar] [CrossRef]
- Ali, Y.B.; Verger, R.; Abousalham, A. Lipases or Esterases: Does It Really Matter? Toward a New Bio-Physico-Chemical Classification. In Lipases and Phospholipases; Humana Press: Totowa, NJ, USA, 2012; pp. 31–51. [Google Scholar]
- Webb, E.C. Enzyme Nomenclature 1992: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes; Academic Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Chahinian, H.; Sarda, L. Distinction between esterases and lipases: Comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 2009, 16, 1149–1161. [Google Scholar] [CrossRef]
- Schmidt, M.; Zahn, S.; Gehlhaar, F.; Prager, A.; Griebel, J.; Kahnt, A.; Knolle, W.; Konieczny, R.; Glaser, R.; Schulze, A. Radiation-Induced Graft Immobilization (RIGI): Covalent Binding of Non-Vinyl Compounds on Polymer Membranes. Polymers 2021, 13, 1849. [Google Scholar] [CrossRef]
- Jahangiri, E.; Thomas, I.; Schulze, A.; Seiwert, B.; Cabana, H.; Schlosser, D. Characterisation of electron beam irradiation-immobilised laccase for application in wastewater treatment. Sci. Total Environ. 2018, 624, 309–322. [Google Scholar] [CrossRef]
- Luick, J.R.; Mazrimas, J.A. Biological Effects of Ionizing Radiation on Milk Synthesis. III. Effects on Milk Lipase, Esterase, Alkaline Phosphatase, and Lactoperoxidase Activities. J. Dairy Sci. 1966, 49, 1500–1504. [Google Scholar] [CrossRef]
- Bobrowski, K. Radiation chemistry of liquid systems. In Applications of Ionizing Radiation in Materials Processing; Sun, Y., Chmielewski, A.G., Eds.; Institute of Nuclear Chemistry and Technology: Warszawa, Poland, 2017. [Google Scholar]
- Adem, E.; Burillo, G.; Muñoz, E.; Rickards, J.; Cota, L.; Avalos-Borja, M. Electron and proton irradiation of poly(vinylidene fluoride): Characterization by electron paramagnetic resonance. Polym. Degrad. Stab. 2003, 81, 75–79. [Google Scholar] [CrossRef]
- Forsythe, J.S.; Hill, D.J.T. The radiation chemistry of fluoropolymers. Prog. Polym. Sci. 2000, 25, 101–136. [Google Scholar] [CrossRef]
- Buxton, G.V.; Greenstock, C.L.; Helman, W.P.; Ross, A.B. Critical Review of rate constants for reactions of hydrated electrons, hydrogen atoms and hydroxyl radicals OH/O− in Aqueous Solution. J. Phys. Chem. Ref. Data 1988, 17, 513–886. [Google Scholar] [CrossRef] [Green Version]
- Spinks, J.W.T.; Woods, R.J. An Introduction to Radiation Chemistry, 3rd ed.; Wiley: New York, NJ, USA; Chichester, UK, 1990. [Google Scholar]
- Plant, A.G.; Kos, B.; Jazbec, A.; Snoj, L.; Najdanovic-Visak, V.; Joyce, M.J. Nuclear-driven production of renewable fuel additives from waste organics. Commun. Chem. 2021, 4, 132. [Google Scholar] [CrossRef]
- Zdarta, J.; Jesionowski, T.; Pinelo, M.; Meyer, A.S.; Iqbal, H.M.N.; Bilal, M.; Nguyen, L.N.; Nghiem, L.D. Free and immobilized biocatalysts for removing micropollutants from water and wastewater: Recent progress and challenges. Bioresour. Technol. 2022, 344, 126201. [Google Scholar] [CrossRef]
- Syed, Q.A.; Hassan, A.; Sharif, S.; Ishaq, A.; Saeed, F.; Afzaal, M.; Hussain, M.; Anjum, F.M. Structural and functional properties of milk proteins as affected by heating, high pressure, Gamma and ultraviolet irradiation: A review. Int. J. Food Prop. 2021, 24, 871–884. [Google Scholar] [CrossRef]
- Lalande, M.; Schwob, L.; Vizcaino, V.; Chirot, F.; Dugourd, P.; Schlatholter, T.; Poully, J.C. Direct Radiation Effects on the Structure and Stability of Collagen and Other Proteins. Chembiochem 2019, 20, 2972–2980. [Google Scholar] [CrossRef] [Green Version]
- Cavalcante, F.T.T.; Cavalcante, A.L.G.; Sousa, I.G.D.; Neto, F.S.; Santos, J.C.S.D. Current Status and Future Perspectives of Supports and Protocols for Enzyme Immobilization. Catalysts 2021, 11, 1222. [Google Scholar] [CrossRef]
- Gupta, R.; Rathi, P.; Gupta, N.; Bradoo, S. Lipase assays for conventional and molecular screening: An overview. Biotechnol. Appl. Biochem. 2003, 37, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Noble, J.E.; Knight, A.E.; Reason, A.J.; Matola, A.D.; Bailey, M.J. A comparison of protein quantitation assays for biopharmaceutical applications. Mol. Biotechnol. 2007, 37, 99–111. [Google Scholar] [CrossRef]
- Zimmermann, R.; Dukhin, S.; Werner, C. Electrokinetic Measurements Reveal Interfacial Charge at Polymer Films Caused by Simple Electrolyte Ions. J. Phys. Chem. B 2001, 105, 8544–8549. [Google Scholar] [CrossRef]
- Pinem, J.A.; Wardani, A.K.; Aryanti, P.T.P.; Khoiruddin, K.; Wenten, I.G. Hydrophilic Modification of Polymeric Membrane using Graft Polymerization Method: A Mini Review. IOP Conf. Ser. Mater. Sci. Eng. 2019, 547, 012054. [Google Scholar] [CrossRef]
- Rezakazemi, M.; Dashti, A.; Harami, H.R.; Hajilari, N.; Inamuddin. Fouling-resistant membranes for water reuse. Environ. Chem. Lett. 2018, 16, 715–763. [Google Scholar] [CrossRef]
- Xu, F.; Wang, W.H.; Tan, Y.J.; Bruening, M.L. Facile trypsin immobilization in polymeric membranes for rapid, efficient protein digestion. Anal. Chem. 2010, 82, 10045–10051. [Google Scholar] [CrossRef] [Green Version]
- Gul, A.; Hruza, J.; Yalcinkaya, F. Fouling and Chemical Cleaning of Microfiltration Membranes: A Mini-Review. Polymers 2021, 13, 846. [Google Scholar] [CrossRef]
- Díez, B.; Rosal, R. A critical review of membrane modification techniques for fouling and biofouling control in pressure-driven membrane processes. Nanotechnol. Environ. Eng. 2020, 5, 1–21. [Google Scholar] [CrossRef]
- Hermansyah, H.; Wijanarko, A.; Kubo, M.; Shibasaki-Kitakawa, N.; Yonemoto, T. Rigorous kinetic model considering positional specificity of lipase for enzymatic stepwise hydrolysis of triolein in biphasic oil-water system. Bioprocess Biosyst. Eng. 2010, 33, 787–796. [Google Scholar] [CrossRef]
- Alongi, M.; Lucci, P.; Clodoveo, M.L.; Schena, F.P.; Calligaris, S. Oleogelation of extra virgin olive oil by different oleogelators affects the physical properties and the stability of bioactive compounds. Food Chem. 2022, 368, 130779. [Google Scholar] [CrossRef]
- Li, J.; Guo, R.; Bi, Y.; Zhang, H.; Xu, X. Comprehensive evaluation of saturated monoglycerides for the forming of oleogels. LWT 2021, 151, 112061. [Google Scholar] [CrossRef]
- Monie, A.; Franceschi, S.; Balayssac, S.; Malet-Martino, M.; Delample, M.; Perez, E.; Garrigues, J.C. Study of rapeseed oil gelation induced by commercial monoglycerides using a chemometric approach. Food Chem. 2022, 369, 130870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, D.; Tian, Y.; Li, H.; Gong, T.; Luo, J.; Ruan, J.; Gong, T.; Zhang, Z. Comparison of three in-situ gels composed of different oil types. Int. J. Pharm. 2020, 587, 119707. [Google Scholar] [CrossRef] [PubMed]
- Uncu, O.; Ozen, B.; Tokatli, F. Use of FTIR and UV-visible spectroscopy in determination of chemical characteristics of olive oils. Talanta 2019, 201, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Crizel, R.L.; Hoffmann, J.F.; Zandoná, G.P.; Lobo, P.M.S.; Jorge, R.O.; Chaves, F.C. Characterization of Extra Virgin Olive Oil from Southern Brazil. Eur. J. Lipid Sci. Technol. 2020, 122, 1900347. [Google Scholar] [CrossRef]
- Bialecka-Florjanczyk, E.; Fabiszewska, A.U.; Krzyczkowska, J.; Kurylowicz, A. Synthetic and Natural Lipase Inhibitors. Mini Rev. Med. Chem. 2018, 18, 672–683. [Google Scholar] [CrossRef]





| Factor | Name | Units | Type | Lower Limit | Upper Limit |
|---|---|---|---|---|---|
| A | enzyme concentration | g L−1 | numeric | 1.0 | 10.0 |
| B | impregnation time | min | numeric | 0.1 | 10.0 |
| C | irradiation dose | kGy | numeric | 50 | 200 |
| D | enzyme | – | categoric | FE-01 | EL-01 |
| Source | Sum of Squares | Df | Mean Square | F-Value | p-Value |
|---|---|---|---|---|---|
| Block | 0.067 | 1 | 0.067 | - | - |
| Model | 76.208 | 17 | 4.483 | 41.24 | <0.0001 |
| A: enzyme conc. | 16.805 | 1 | 16.805 | 154.59 | <0.0001 |
| B: impreg. time | 0.106 | 1 | 0.106 | 0.98 | 0.3314 |
| C: irrad. dose | 8.632 | 1 | 8.632 | 79.41 | <0.0001 |
| D: enzyme | 0.172 | 1 | 0.172 | 1.58 | 0.2184 |
| AB | 0.028 | 1 | 0.028 | 0.26 | 0.6130 |
| AC | 4.305 | 1 | 4.305 | 39.60 | <0.0001 |
| AD | 0.500 | 1 | 0.500 | 4.60 | 0.0405 |
| BC | 0.053 | 1 | 0.053 | 0.49 | 0.4891 |
| CD | 0.897 | 1 | 0.897 | 8.25 | 0.0075 |
| A² | 2.289 | 1 | 2.289 | 21.06 | <0.0001 |
| B² | 1.667 | 1 | 1.667 | 15.33 | 0.0005 |
| C² | 3.678 | 1 | 3.678 | 33.84 | <0.0001 |
| A²B | 1.675 | 1 | 1.675 | 15.40 | 0.0005 |
| AB² | 1.096 | 1 | 1.096 | 10.08 | 0.0035 |
| B²C | 0.683 | 1 | 0.683 | 6.28 | 0.0181 |
| B³ | 1.190 | 1 | 1.190 | 10.95 | 0.0025 |
| C³ | 0.766 | 1 | 0.766 | 7.04 | 0.0128 |
| Residual | 3.152 | 29 | 0.109 | - | - |
| Lack-of-Fit (LOF) | 2.735 | 21 | 0.130 | 2.50 | 0.0922 |
| Pure Error | 0.417 | 8 | 0.052 | - | - |
| Cor Total | 79.428 | 47 | - | - | - |
| Vmax/µM s−1 | Km/µM | V/K/10−3 s−1 | Asp/U m−2 |
|---|---|---|---|
| 2.7 ± 0.4 | 62.9 ± 19.3 | 44.1 ± 7.7 | 823 ± 118 |
| Sample | Elemental Composition/at % | Elemental Ratio/% | |||||||
|---|---|---|---|---|---|---|---|---|---|
| C | F | O | N | S | Si | F/C | O/C | N/C | |
| PVDF-Ref | 59.18 | 37.21 | 3.13 | 0.00 | 0.00 | 0.49 | 62.89 | 5.29 | 0.00 |
| ±0.57 | ±1.05 | ±0.40 | ±0.00 | ±0.00 | ±0.10 | ±2.38 | ±0.62 | ±0.00 | |
| PVDF-g-EL | 61.51 | 32.27 | 5.03 | 0.58 | 0.00 | 0.62 | 52.47 | 8.17 | 0.93 |
| ±0.47 | ±1.32 | ±0.67 | ±0.10 | ±0.00 | ±0.36 | ±2.50 | ±1.04 | ±0.15 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, M.; Prager, A.; Schönherr, N.; Gläser, R.; Schulze, A. Reagent-Free Immobilization of Industrial Lipases to Develop Lipolytic Membranes with Self-Cleaning Surfaces. Membranes 2022, 12, 599. https://doi.org/10.3390/membranes12060599
Schmidt M, Prager A, Schönherr N, Gläser R, Schulze A. Reagent-Free Immobilization of Industrial Lipases to Develop Lipolytic Membranes with Self-Cleaning Surfaces. Membranes. 2022; 12(6):599. https://doi.org/10.3390/membranes12060599
Chicago/Turabian StyleSchmidt, Martin, Andrea Prager, Nadja Schönherr, Roger Gläser, and Agnes Schulze. 2022. "Reagent-Free Immobilization of Industrial Lipases to Develop Lipolytic Membranes with Self-Cleaning Surfaces" Membranes 12, no. 6: 599. https://doi.org/10.3390/membranes12060599
APA StyleSchmidt, M., Prager, A., Schönherr, N., Gläser, R., & Schulze, A. (2022). Reagent-Free Immobilization of Industrial Lipases to Develop Lipolytic Membranes with Self-Cleaning Surfaces. Membranes, 12(6), 599. https://doi.org/10.3390/membranes12060599