Supporting the Diagnosis of Fabry Disease Using a Natural Language Processing-Based Approach
Abstract
:1. Introduction
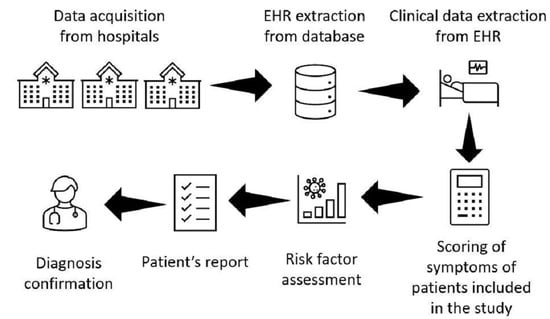
2. Materials and Methods
2.1. Studied Population
2.2. FD Risk Factor Development
2.3. Natural Language Processing
2.4. FD Risk Score Implementation
2.5. Statistical Analysis
3. Results
Patients with Diagnosed FD
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehta, A.; Beck, M.; Eyskens, F.; Feliciani, C.; Kantola, I.; Ramaswami, U.; Rolfs, A.; Rivera, A.; Waldek, S.; Germain, D. Fabry disease: A review of current management strategies. QJM Int. J. Med. 2010, 103, 641–659. [Google Scholar] [CrossRef] [PubMed]
- Garman, S.C.; Garboczi, D.N. The molecular defect leading to Fabry disease: Structure of human α-galactosidase. J. Mol. Biol. 2004, 337, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Aerts, J.M.; Groener, J.E.; Kuiper, S.; Donker-Koopman, W.E.; Strijland, A.; Ottenhoff, R.; van Roomen, C.; Mirzaian, M.; Wijburg, F.A.; Linthorst, G.E.; et al. Elevated globotriaosylsphingosine is a hallmark of Fabry disease. Proc. Natl. Acad. Sci. USA 2008, 105, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Hwu, W.L.; Chien, Y.H.; Lee, N.C.; Chiang, S.C.; Dobrovolny, R.; Huang, A.C.; Yeh, H.Y.; Chao, M.C.; Lin, S.J.; Kitagawa, T.; et al. Newborn screening for Fabry disease in Taiwan reveals a high incidence of the later-onset GLA mutation c. 936+ 919G> A (IVS4+ 919G> A). Hum. Mutat. 2009, 30, 1397–1405. [Google Scholar] [CrossRef]
- MacDermot, K.; Holmes, A.; Miners, A. Anderson-Fabry disease: Clinical manifestations and impact of disease in a cohort of 98 hemizygous males. J. Med. Genet. 2001, 38, 750–760. [Google Scholar] [CrossRef]
- Mechtler, T.P.; Stary, S.; Metz, T.F.; De Jesus, V.R.; Greber-Platzer, S.; Pollak, A.; Herkner, K.R.; Streubel, B.; Kasper, D.C. Neonatal screening for lysosomal storage disorders: Feasibility and incidence from a nationwide study in Austria. Lancet 2012, 379, 335–341. [Google Scholar] [CrossRef]
- Spada, M.; Pagliardini, S.; Yasuda, M.; Tukel, T.; Thiagarajan, G.; Sakuraba, H.; Ponzone, A.; Desnick, R.J. High incidence of later-onset Fabry disease revealed by newborn screening. Am. J. Hum. Genet. 2006, 79, 31–40. [Google Scholar] [CrossRef]
- Di Risi, T.; Vinciguerra, R.; Cuomo, M.; Della Monica, R.; Riccio, E.; Cocozza, S.; Imbriaco, M.; Duro, G.; Pisani, A.; Chiariotti, L. DNA methylation impact on Fabry disease. Clin. Epigenetics 2021, 13, 24. [Google Scholar] [CrossRef]
- Echevarria, L.; Benistan, K.; Toussaint, A.; Dubourg, O.; Hagege, A.; Eladari, D.; Jabbour, F.; Beldjord, C.; De Mazancourt, P.; Germain, D. X-chromosome inactivation in female patients with Fabry disease. Clin. Genet. 2016, 89, 44–54. [Google Scholar] [CrossRef]
- Germain, D.P.; Oliveira, J.P.; Bichet, D.G.; Yoo, H.W.; Hopkin, R.J.; Lemay, R.; Politei, J.; Wanner, C.; Wilcox, W.R.; Warnock, D.G. Use of a rare disease registry for establishing phenotypic classification of previously unassigned GLA variants: A consensus lassification system by a multispecialty Fabry disease genotype–phenotype workgroup. J. Med. Genet. 2020, 57, 542–551. [Google Scholar] [CrossRef]
- Politei, J.M.; Bouhassira, D.; Germain, D.P.; Goizet, C.; Guerrero-Sola, A.; Hilz, M.J.; Hutton, E.J.; Karaa, A.; Liguori, R.; Üçeyler, N.; et al. Pain in Fabry disease: Practical recommendations for diagnosis and treatment. CNS Neurosci. Ther. 2016, 22, 568–576. [Google Scholar] [CrossRef] [PubMed]
- MacDermot, K.; Holmes, A.; Miners, A. Natural history of Fabry disease in affected males and obligate carrier females. J. Inherit. Metab. Dis. 2001, 24, 13. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Ricci, R.; Widmer, U.; Dehout, F.; Garcia de Lorenzo, A.; Kampmann, C.; Linhart, A.; Sunder-Plassmann, G.; Ries, M.; Beck, M. Fabry disease defined: Baseline clinical manifestations of 366 patients in the Fabry Outcome Survey. Eur. J. Clin. Investig. 2004, 34, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Reisin, R.; Perrin, A.; García-Pavía, P. Time delays in the diagnosis and treatment of Fabry disease. Int. J. Clin. Pract. 2017, 71, e12914. [Google Scholar] [CrossRef]
- Rosa Neto, N.S.; de Barros Bento, J.C.; Pereira, R.M.R. Higher rate of rheumatic manifestations and delay in diagnosis in Brazilian Fabry disease patients. Adv. Rheumatol. 2020, 60, 7. [Google Scholar] [CrossRef]
- Thomas, A.S.; Mehta, A.B. Difficulties and barriers in diagnosing Fabry disease: What can be learnt from the literature? Expert Opin. Med. Diagn. 2013, 7, 589–599. [Google Scholar] [CrossRef]
- Nestler-Parr, S.; Korchagina, D.; Toumi, M.; Pashos, C.L.; Blanchette, C.; Molsen, E.; Morel, T.; Simoens, S.; Kaló, Z.; Gatermann, R.; et al. Challenges in research and health technology assessment of rare disease technologies: Report of the ISPOR rare disease special interest group. Value Health 2018, 21, 493–500. [Google Scholar] [CrossRef]
- Nowicki, M.; Bazan-Socha, S.; Błazejewska-Hyzorek, B.; Gellert, R.; Imiela, J.; Ka´zmierczak, J.; Kłopotowski, M.; Oko-Sarnowska, Z.; Pawlaczyk, K.; Ponikowski, P.; et al. Enzyme replacement therapy in Fabry disease in Poland: Position statement. Pol. Arch. Intern. Med. 2020, 130, 91–97. [Google Scholar] [CrossRef]
- Germain, D.P.; Charrow, J.; Desnick, R.J.; Guffon, N.; Kempf, J.; Lachmann, R.H.; Lemay, R.; Linthorst, G.E.; Packman, S.; Scott, C.R.; et al. Ten-year outcome of enzyme replacement therapy with agalsidase beta in patients with Fabry disease. J. Med. Genet. 2015, 52, 353–358. [Google Scholar] [CrossRef]
- Rombach, S.M.; Smid, B.E.; Bouwman, M.G.; Linthorst, G.E.; Dijkgraaf, M.G.; Hollak, C.E. Long term enzyme replacement therapy for Fabry disease: Effectiveness on kidney, heart and brain. Orphanet J. Rare Dis. 2013, 8, 47. [Google Scholar] [CrossRef]
- Weidemann, F.; Sanchez-Niño, M.D.; Politei, J.; Oliveira, J.P.; Wanner, C.; Warnock, D.G.; Ortiz, A. Fibrosis: A key feature of Fabry disease with potential therapeutic implications. Orphanet J. Rare Dis. 2013, 8, 116. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.A.; Malmenäs, M.; Deegan, P.B.; Elliott, P.M.; Ginsberg, L.; Hajioff, D.; Ioannidis, A.S.; Orteu, C.H.; Ramaswami, U.; West, M.; et al. Fabry International Prognostic Index: A predictive severity score for Anderson-Fabry disease. J. Med. Genet. 2012, 49, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Jefferies, J.L.; Spencer, A.K.; Lau, H.A.; Nelson, M.W.; Giuliano, J.D.; Zabinski, J.W.; Boussios, C.; Curhan, G.; Gliklich, R.E.; Warnock, D.G. A new approach to identifying patients with elevated risk for Fabry disease using a machine learning algorithm. Orphanet J. Rare Dis. 2021, 16, 518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Abu-el Rub, N.; Gray, J.; Pham, H.A.; Zhou, Y.; Manion, F.J.; Liu, M.; Song, X.; Xu, H.; Rouhizadeh, M.; et al. COVID-19 SignSym: A fast adaptation of a general clinical NLP tool to identify and normalize COVID-19 signs and symptoms to OMOP common data model. J. Am. Med. Inform. Assoc. 2021, 28, 1275–1283. [Google Scholar] [CrossRef] [PubMed]
- Neuraz, A.; Lerner, I.; Digan, W.; Paris, N.; Tsopra, R.; Rogier, A.; Baudoin, D.; Cohen, K.B.; Burgun, A.; Garcelon, N.; et al. Natural language processing for rapid response to emergent diseases: Case study of calcium channel blockers and hypertension in the COVID-19 pandemic. J. Med. Internet Res. 2020, 22, e20773. [Google Scholar] [CrossRef] [PubMed]
- Lybarger, K.; Ostendorf, M.; Thompson, M.; Yetisgen, M. Extracting COVID-19 diagnoses and symptoms from clinical text: A new annotated corpus and neural event extraction framework. J. Biomed. Inform. 2021, 117, 103761. [Google Scholar] [CrossRef]
- Chapman, A.; Peterson, K.; Turano, A.; Box, T.; Wallace, K.; Jones, M. A Natural Language Processing System for National COVID-19 Surveillance in the US Department of Veterans Affairs. In Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, Online, 9 July 2020; Association for Computational Linguistics: Toronto, ON, Canada, 2020. [Google Scholar]
- Schöning, V.; Liakoni, E.; Drewe, J.; Hammann, F. Automatic identification of risk factors for SARS-CoV-2 positivity and severe linical outcomes of COVID-19 using Data Mining and Natural Language Processing. medRxiv 2021. [Google Scholar] [CrossRef]
- Oyebode, O.; Ndulue, C.; Adib, A.; Mulchandani, D.; Suruliraj, B.; Orji, F.A.; Chambers, C.T.; Meier, S.; Orji, R. Health, psychosocial, and social issues emanating from the COVID-19 pandemic based on social media comments: Text mining and thematic analysis approach. JMIR Med. Inform. 2021, 9, e22734. [Google Scholar] [CrossRef]
- Honnibal, M.; Montani, I.; Van Landeghem, S.; Boyd, A. spaCy: Industrial-Strength Natural Language Processing in Python. Available online: https://github.com/explosion/spaCy (accessed on 6 January 2022).
- Kluyver, T.; Ragan-Kelley, B.; Pérez, F.; Granger, B.; Bussonnier, M.; Frederic, J.; Kelley, K.; Hamrick, J.; Grout, J.; Corlay, S.; et al. Jupyter Notebooks—A publishing format for reproducible computational workflows. In Proceedings of the Positioning and Power in Academic Publishing: Players, Agents and Agendas, Göttingen, Germany, 7–9 June 2016; Loizides, F., Schmidt, B., Eds.; IOS Press: Amsterdam, The Netherlands, 2016; pp. 87–90. [Google Scholar]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Pandas Development Team. pandas-dev/pandas: Pandas. Available online: https://github.com/pandas-dev/pandas (accessed on 6 January 2022).
- Ortiz, A.; Germain, D.P.; Desnick, R.J.; Politei, J.; Mauer, M.; Burlina, A.; Eng, C.; Hopkin, R.J.; Laney, D.; Linhart, A.; et al. Fabry disease revisited: Management and treatment recommendations for adult patients. Mol. Genet. Metab. 2018, 123, 416–427. [Google Scholar] [CrossRef]
- Tuttolomondo, A.; Pecoraro, R.; Simonetta, I.; Miceli, S.; Pinto, A.; Licata, G. Anderson-Fabry disease: A multiorgan disease. Curr. Pharm. Des. 2013, 19, 5974–5996. [Google Scholar] [CrossRef] [PubMed]
- Delarosa-Rodríguez, R.; Santotoribio, J.D.; Paula, H.A.; González-Meneses, A.; García-Morillo, S.; Jiménez-Arriscado, P.; Guerrero, J.M.; Macher, H.C. Accuracy diagnosis improvement of Fabry disease from dried blood spots: Enzyme activity, lyso-Gb3 accumulation and GLA gene sequencing. Clin. Genet. 2021, 99, 761–771. [Google Scholar] [CrossRef] [PubMed]
- Scalia, S. DBS assay in the diagnosis of Fabry disease. Giornale Di Tecniche Nefrologiche E Dialitiche 2017, 29, S5–S6. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M.L. seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Faviez, C.; Chen, X.; Garcelon, N.; Neuraz, A.; Knebelmann, B.; Salomon, R.; Lyonnet, S.; Saunier, S.; Burgun, A. Diagnosis support systems for rare diseases: A scoping review. Orphanet J. Rare Dis. 2020, 15, 94. [Google Scholar] [CrossRef]
- Schaefer, J.; Lehne, M.; Schepers, J.; Prasser, F.; Thun, S. The use of machine learning in rare diseases: A scoping review. Orphanet J. Rare Dis. 2020, 15, 145. [Google Scholar] [CrossRef]
- Lee, S.N.; Kim, G.H.; Yoo, K.D. A Case of a 50-Year-Old Woman with Typical Fabry Disease Who Showed Serial Electrocardio- graphic and Echocardiographic Changes over a 17-Year Period. Case Rep. Cardiol. 2019, 2019, 9385361. [Google Scholar]
- Lidove, O.; Barbey, F.; Niu, D.M.; Brand, E.; Nicholls, K.; Bizjajeva, S.; Hughes, D.A. Fabry in the older patient: Clinical consequences and possibilities for treatment. Mol. Genet. Metab. 2016, 118, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Radford, A.; Narasimhan, K.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://openai.com/research/language-unsupervised (accessed on 6 January 2022).
- Rasmy, L.; Xiang, Y.; Xie, Z.; Tao, C.; Zhi, D. Med-BERT: Pretrained contextualized embeddings on large-scale structured electronic health records for disease prediction. NPJ Digit. Med. 2021, 4, 86. [Google Scholar] [CrossRef] [PubMed]
- Mroczkowski, R.; Rybak, P.; Wróblewska, A.; Gawlik, I. HerBERT: Efficiently pretrained transformer-based language model for Polish. arXiv 2021, arXiv:2105.01735. [Google Scholar]






| Study Group (n = 13) | Control Group (n = 19,372) | p-Value | |
|---|---|---|---|
| Sex (% female) | 38.5% | 50.5% | 0.38 |
| Mean age (SD) | 45.2 (10.5) | 55.5 (13.3) | p < 0.05 |
| Cardiovascular diseases (%) | 10 (76.9%) | 6641 (34.3%) | p < 0.05 |
| Skin changes (%) | 8 (61.5%) | 149 (0.8%) | p < 0.05 |
| Neurological disorders (%) | 7 (53.8%) | 5034 (26.0%) | p < 0.05 |
| Kidney diseases (%) | 4 (30.8%) | 2030 (10.5%) | p < 0.05 |
| Eye disorder (%) | 0 (0%) | 298 (1.5%) | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michalski, A.A.; Lis, K.; Stankiewicz, J.; Kloska, S.M.; Sycz, A.; Dudziński, M.; Muras-Szwedziak, K.; Nowicki, M.; Bazan-Socha, S.; Dabrowski, M.J.; et al. Supporting the Diagnosis of Fabry Disease Using a Natural Language Processing-Based Approach. J. Clin. Med. 2023, 12, 3599. https://doi.org/10.3390/jcm12103599
Michalski AA, Lis K, Stankiewicz J, Kloska SM, Sycz A, Dudziński M, Muras-Szwedziak K, Nowicki M, Bazan-Socha S, Dabrowski MJ, et al. Supporting the Diagnosis of Fabry Disease Using a Natural Language Processing-Based Approach. Journal of Clinical Medicine. 2023; 12(10):3599. https://doi.org/10.3390/jcm12103599
Chicago/Turabian StyleMichalski, Adrian A., Karol Lis, Joanna Stankiewicz, Sylwester M. Kloska, Arkadiusz Sycz, Marek Dudziński, Katarzyna Muras-Szwedziak, Michał Nowicki, Stanisława Bazan-Socha, Michal J. Dabrowski, and et al. 2023. "Supporting the Diagnosis of Fabry Disease Using a Natural Language Processing-Based Approach" Journal of Clinical Medicine 12, no. 10: 3599. https://doi.org/10.3390/jcm12103599
APA StyleMichalski, A. A., Lis, K., Stankiewicz, J., Kloska, S. M., Sycz, A., Dudziński, M., Muras-Szwedziak, K., Nowicki, M., Bazan-Socha, S., Dabrowski, M. J., & Basak, G. W. (2023). Supporting the Diagnosis of Fabry Disease Using a Natural Language Processing-Based Approach. Journal of Clinical Medicine, 12(10), 3599. https://doi.org/10.3390/jcm12103599