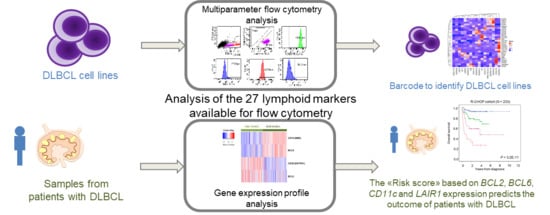
Phenotypic Characterization of Diffuse Large B-Cell Lymphoma Cells and Prognostic Impact
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Flow Cytometry Analysis
2.3. Cell Viability Assay
2.4. Gene Expression Profiling and Statistical Analyses
2.5. Building the B-Cell Marker Risk Score
2.6. Interaction Effect Quantification
3. Results
3.1. A Barcode to Identify DLBCL-Derived Cell Lines by MFC
3.2. CD39 is A Useful Marker to Discriminate Between ABC and GCB DLBCL Tumor Samples
3.3. The B Cell Marker Risk Score
3.4. BCL6 Protein Expression is Correlated with DLBCL Cell Response to Etoposide
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DLBCL | Diffuse large B-cell lymphoma |
| GEP | Gene expression profiling |
| COO | Cell of origin |
| GCB | Germinal-center B-cell–like subgroup |
| ABC | Activated B cell–like subtype |
| R | Rituximab |
| CHOP | Cyclophosphamide, doxorubicin, vincristine, and prednisone |
| EFS | Event-free survival |
| OS | Overall survival |
| FCM | Flow cytometry |
| FSC | Forward scatter |
| SSC | Side scatter |
| NF-kB | Nuclear factor kB |
| ERK | Extracellular signal-regulated kinases |
| IC50 | Inhibitory concentration 50 |
| INPP5D | Src homology 2 domain containing inositol polyphosphate 5-phosphatase 1 |
| IHC | Immunohistochemistry |
| PCR | Polymerase chain reaction |
| PTPN6 | Protein Tyrosine Phosphatase Non-Receptor Type 6 |
| SAM | Significance analysis of microarray |
| STR | Short tandem repeat |
| SYK | Spleen tyrosine kinase |
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics, 2012. CA. Cancer J. Clin. 2012, 62, 10–29. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Staudt, L.M.; Wilson, W.H. Diffuse large B-cell lymphoma-treatment approaches in the molecular era. Nat. Rev. Clin. Oncol. 2014, 11, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Veldman-Jones, M.H.; Lai, Z.; Wappett, M.; Harbron, C.G.; Barrett, J.C.; Harrington, E.A.; Thress, K.S. Reproducible, Quantitative, and Flexible Molecular Subtyping of Clinical DLBCL Samples Using the NanoString nCounter System. Clin. Cancer Res. 2015, 21, 2367–2378. [Google Scholar] [CrossRef] [PubMed]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Müller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S.; et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Wright, G.W.; Williams, P.M.; Lih, C.-J.; Walsh, W.; Jaffe, E.S.; Rosenwald, A.; Campo, E.; Chan, W.C.; Connors, J.M.; et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin embedded tissue. Blood 2014, 123, 1214–1217. [Google Scholar] [CrossRef] [PubMed]
- Moreaux, J.; Klein, B.; Bataille, R.; Descamps, G.; Maïga, S.; Hose, D.; Goldschmidt, H.; Jauch, A.; Rème, T.; Jourdan, M.; et al. A high-risk signature for patients with multiple myeloma established from the molecular classification of human myeloma cell lines. Haematologica 2011, 96, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Bou Samra, E.; Klein, B.; Commes, T.; Moreaux, J. Development of gene expression-based risk score in cytogenetically normal acute myeloid leukemia patients. Oncotarget 2012, 3, 824–832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bret, C.; Klein, B.; Moreaux, J. Gene expression-based risk score in diffuse large B-cell lymphoma. Oncotarget 2012, 3, 1700–1710. [Google Scholar] [CrossRef] [Green Version]
- van Dongen, J.J.M.; Lhermitte, L.; Böttcher, S.; Almeida, J.; van der Velden, V.H.J.; Flores-Montero, J.; Rawstron, A.; Asnafi, V.; Lécrevisse, Q.; Lucio, P.; et al. EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012, 26, 1908–1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Dongen, J.J.M.; Orfao, A. EuroFlow: Resetting leukemia and lymphoma immunophenotyping. Basis for companion diagnostics and personalized medicine. Leukemia 2012, 26, 1899–1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alaterre, E.; Raimbault, S.; Garcia, J.-M.; Rème, T.; Requirand, G.; Klein, B.; Moreaux, J. Automated and simplified identification of normal and abnormal plasma cells in Multiple Myeloma by flow cytometry. Cytom. Part B Clin. Cytom. 2017, 94, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Lenz, G.; Wright, G.; Dave, S.S.; Xiao, W.; Powell, J.; Zhao, H.; Xu, W.; Tan, B.; Goldschmidt, N.; Iqbal, J.; et al. Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar] [CrossRef] [PubMed]
- Shaknovich, R.; Geng, H.; Johnson, N.A.; Tsikitas, L.; Cerchietti, L.; Greally, J.M.; Gascoyne, R.D.; Elemento, O.; Melnick, A. DNA methylation signatures define molecular subtypes of diffuse large B-cell lymphoma. Blood 2010, 116, e81–e89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dybkær, K.; Bøgsted, M.; Falgreen, S.; Bødker, J.S.; Kjeldsen, M.K.; Schmitz, A.; Bilgrau, A.E.; Xu-Monette, Z.Y.; Li, L.; Bergkvist, K.S.; et al. Diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J. Clin. Oncol. 2015, 33, 1379–1388. [Google Scholar] [CrossRef]
- Kassambara, A.; Rème, T.; Jourdan, M.; Fest, T.; Hose, D.; Tarte, K.; Klein, B. GenomicScape: An easy-to-use web tool for gene expression data analysis. Application to investigate the molecular events in the differentiation of B cells into plasma cells. PLoS Comput. Biol. 2015, 11, e1004077. [Google Scholar] [CrossRef]
- Kassambara, A.; Hose, D.; Moreaux, J.; Walker, B.A.; Protopopov, A.; Reme, T.; Pellestor, F.; Pantesco, V.; Jauch, A.; Morgan, G.; et al. Genes with a spike expression are clustered in chromosome (sub)bands and spike (sub)bands have a powerful prognostic value in patients with multiple myeloma. Haematologica 2012, 97, 622–630. [Google Scholar] [CrossRef]
- Hothorn, T.; Lausen, B. On the exact distribution of maximally selected rank statistics. Stat. Med. 2012, 3178–3191. [Google Scholar] [CrossRef]
- Alaterre, E.; Raimbault, S.; Goldschmidt, H.; Bouhya, S.; Requirand, G.; Robert, N.; Boireau, S.; Seckinger, A.; Hose, D.; Klein, B.; et al. CD24, CD27, CD36 and CD302 gene expression for outcome prediction in patients with multiple myeloma. Oncotarget 2017, 8, 98931–98944. [Google Scholar] [CrossRef]
- Greco, W.R.; Bravo, G.; Parsons, J.C. The search for synergy: A critical review from a response surface perspective. Pharmacol. Rev. 1995, 47, 331–385. [Google Scholar] [PubMed]
- Combes, E.; Andrade, A.F.; Tosi, D.; Michaud, H.-A.; Coquel, F.; Garambois, V.; Desigaud, D.; Jarlier, M.; Coquelle, A.; Pasero, P.; et al. Inhibition of Ataxia-Telangiectasia Mutated and RAD3-related (ATR) overcomes oxaliplatin resistance and promotes anti-tumor immunity in colorectal cancer. Cancer Res. 2019, 79, 2933–2946. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, A.L.; Young, R.M.; Staudt, L.M. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 2012, 30, 565–610. [Google Scholar] [CrossRef] [PubMed]
- Deeb, S.J.; D’Souza, R.C.J.; Cox, J.; Schmidt-Supprian, M.; Mann, M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteom. MCP 2012, 11, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Lausen, B.; Schumacher, M. Maximally Selected Rank Statistics. Biometrics 1992, 48, 73–85. [Google Scholar] [CrossRef]
- Pasqualucci, L.; Compagno, M.; Lim, W.K.; Grunn, A.; Nandula, S.V.; Scandurra, M.; Bertoni, F.; Ponzoni, M.; Califano, A.; Bhagat, G.; et al. Mutations in Multiple Genes Cause Deregulation of the NFkB Pathway in Diffuse Large B-Cell Lymphoma. Blood 2008, 112, 801. [Google Scholar]
- Shen, Y.; Iqbal, J.; Huang, J.Z.; Zhou, G.; Chan, W.C. BCL2 protein expression parallels its mRNA level in normal and malignant B cells. Blood 2004, 104, 2936–2939. [Google Scholar] [CrossRef]
- Kjeldsen, M.K.; Perez-Andres, M.; Schmitz, A.; Johansen, P.; Boegsted, M.; Nyegaard, M.; Gaihede, M.; Bukh, A.; Johnsen, H.E.; Orfao, A.; et al. Multiparametric flow cytometry for identification and fluorescence activated cell sorting of five distinct B-cell subpopulations in normal tonsil tissue. Am. J. Clin. Pathol. 2011, 136, 960–969. [Google Scholar] [CrossRef]
- Cerchietti, L.C.; Ghetu, A.F.; Zhu, X.; Da Silva, G.F.; Zhong, S.; Matthews, M.; Bunting, K.L.; Polo, J.M.; Farès, C.; Arrowsmith, C.H.; et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 2010, 17, 400–411. [Google Scholar] [CrossRef]
- Masters, J.R. Human cancer cell lines: Fact and fantasy. Nat. Rev. Mol. Cell Biol. 2000, 1, 233–236. [Google Scholar] [CrossRef]
- Reid, Y.A. Characterization and authentication of cancer cell lines: An overview. Methods Mol. Biol. 2011, 731, 35–43. [Google Scholar] [PubMed]
- Maïga, S.; Brosseau, C.; Descamps, G.; Dousset, C.; Gomez-Bougie, P.; Chiron, D.; Ménoret, E.; Kervoelen, C.; Vié, H.; Cesbron, A.; et al. A simple flow cytometry-based barcode for routine authentication of multiple myeloma and mantle cell lymphoma cell lines. Cytom. Part A 2015, 87, 285–288. [Google Scholar] [CrossRef] [PubMed]
- Parson, W.; Kirchebner, R.; Mühlmann, R.; Renner, K.; Kofler, A.; Schmidt, S.; Kofler, R. Cancer cell line identification by short tandem repeat profiling: Power and limitations. FASEB J. 2005, 19, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Robson, S.C.; Sévigny, J.; Zimmermann, H. The E-NTPDase family of ectonucleotidases: Structure function relationships and pathophysiological significance. Purinergic Signal. 2006, 2, 409–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardoso, C.C.; Auat, M.; Santos-Pirath, I.M.; Rudolf-Oliveira, R.C.M.; da Silva, J.P.; Lange, B.G.; Siegel, D.; de Moraes, A.C.R.; Del Moral, J.A.G.; Santos-Silva, M.C. The importance of CD39, CD43, CD81, and CD95 expression for differentiating B cell lymphoma by flow cytometry. Cytom. Part B Clin. Cytom. 2017, 94, 451–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez-García, G.; Cardesa-Salzmann, T.; Climent, F.; González-Barca, E.; Mercadal, S.; Mate, J.L.; Sancho, J.M.; Arenillas, L.; Serrano, S.; Escoda, L.; et al. Gene-expression profiling and not immunophenotypic algorithms predicts prognosis in patients with diffuse large B-cell lymphoma treated with immunochemotherapy. Blood 2011, 117, 4836–4843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilly, H.; da Silva, M.G.; Vitolo, U.; Jack, A.; Meignan, M.; Lopez-Guillermo, A.; Walewski, J.; André, M.; Johnson, P.W.; Pfreundschuh, M.; et al. Diffuse large B-cell lymphoma (DLBCL): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015, 26, v116–v125. [Google Scholar] [CrossRef]
- Wright, G.; Tan, B.; Rosenwald, A.; Hurt, E.H.; Wiestner, A.; Staudt, L.M. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc. Natl. Acad. Sci. USA 2003, 100, 9991–9996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenz, G.; Wright, G.W.; Emre, N.C.T.; Kohlhammer, H.; Dave, S.S.; Davis, R.E.; Carty, S.; Lam, L.T.; Shaffer, A.L.; Xiao, W.; et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 13520–13525. [Google Scholar] [CrossRef] [Green Version]
- Bret, C.; Klein, B.; Cartron, G.; Schved, J.-F.; Constantinou, A.; Pasero, P.; Moreaux, J. DNA repair in diffuse large B-cell lymphoma: A molecular portrait. Br. J. Haematol. 2015, 169, 296–299. [Google Scholar] [CrossRef]
- Schmidlin, H.; Diehl, S.A.; Blom, B. New insights into the regulation of human B-cell differentiation. Trends Immunol. 2009, 30, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsi, E.D.; Yegappan, S. Lymphoma immunophenotyping: A new era in paraffin-section immunohistochemistry. Adv. Anat. Pathol. 2001, 8, 218–239. [Google Scholar] [CrossRef] [PubMed]
- Bilalovic, N.; Blystad, A.K.; Golouh, R.; Nesland, J.M.; Selak, I.; Trinh, D.; Torlakovic, E. Expression of bcl-6 and CD10 protein is associated with longer overall survival and time to treatment failure in follicular lymphoma. Am. J. Clin. Pathol. 2004, 121, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Aukema, S.M.; Siebert, R.; Schuuring, E.; van Imhoff, G.W.; Kluin-Nelemans, H.C.; Boerma, E.-J.; Kluin, P.M. Double-hit B-cell lymphomas. Blood 2011, 117, 2319–2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Young, K.H.; Medeiros, L.J. Diffuse large B-cell lymphoma. Pathology 2018, 50, 74–87. [Google Scholar] [CrossRef]
- Johnson, N.A.; Slack, G.W.; Savage, K.J.; Connors, J.M.; Ben-Neriah, S.; Rogic, S.; Scott, D.W.; Tan, K.L.; Steidl, C.; Sehn, L.H.; et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J. Clin. Oncol. 2012, 30, 3452–3459. [Google Scholar] [CrossRef]
- Hu, S.; Xu-Monette, Z.Y.; Tzankov, A.; Green, T.; Wu, L.; Balasubramanyam, A.; Liu, W.; Visco, C.; Li, Y.; Miranda, R.N.; et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: A report from The International DLBCL Rituximab-CHOP Consortium Program. Blood 2013, 121, 4021–4031. [Google Scholar] [CrossRef]
- Tsuyama, N.; Sakata, S.; Baba, S.; Mishima, Y.; Nishimura, N.; Ueda, K.; Yokoyama, M.; Terui, Y.; Hatake, K.; Kitagawa, M.; et al. BCL2 expression in DLBCL: Reappraisal of immunohistochemistry with new criteria for therapeutic biomarker evaluation. Blood 2017, 130, 489–500. [Google Scholar] [CrossRef]
- Souers, A.J.; Leverson, J.D.; Boghaert, E.R.; Ackler, S.L.; Catron, N.D.; Chen, J.; Dayton, B.D.; Ding, H.; Enschede, S.H.; Fairbrother, W.J.; et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 2013, 19, 202–208. [Google Scholar] [CrossRef]
- Deeks, E.D. Venetoclax: First Global Approval. Drugs 2016, 76, 979–987. [Google Scholar] [CrossRef]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; Mungall, A.J.; An, J.; Goya, R.; Paul, J.E.; Boyle, M.; Woolcock, B.W.; Kuchenbauer, F.; et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Mandelbaum, J.; Bhagat, G.; Tang, H.; Mo, T.; Brahmachary, M.; Shen, Q.; Chadburn, A.; Rajewsky, K.; Tarakhovsky, A.; Pasqualucci, L.; et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell 2010, 18, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Hansen, M.; Berendse, S.; Marafioti, T.; McNamara, C. Does cell-of-origin or MYC, BCL2 or BCL6 translocation status provide prognostic information beyond the International Prognostic Index score in patients with diffuse large B-cell lymphoma treated with rituximab and chemotherapy? A systematic review. Leuk. Lymphoma 2017, 58, 2403–2418. [Google Scholar] [CrossRef]
- Meyaard, L. The inhibitory collagen receptor LAIR-1 (CD305). J. Leukoc. Biol. 2008, 83, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Son, M.; Santiago-Schwarz, F.; Al-Abed, Y.; Diamond, B. C1q limits dendritic cell differentiation and activation by engaging LAIR-1. Proc. Natl. Acad. Sci. USA 2012, 109, E3160–E3167. [Google Scholar] [CrossRef] [PubMed]
- van der Vuurst de Vries, A.R.; Clevers, H.; Logtenberg, T.; Meyaard, L. Leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1) is differentially expressed during human B cell differentiation and inhibits B cell receptor-mediated signaling. Eur. J. Immunol. 1999, 29, 3160–3167. [Google Scholar] [CrossRef]
- Chen, Z.; Shojaee, S.; Buchner, M.; Geng, H.; Lee, J.W.; Klemm, L.; Titz, B.; Graeber, T.G.; Park, E.; Tan, Y.X.; et al. Signalling thresholds and negative B-cell selection in acute lymphoblastic leukaemia. Nature 2015, 521, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Limnander, A.; Depeille, P.; Freedman, T.S.; Liou, J.; Leitges, M.; Kurosaki, T.; Roose, J.P.; Weiss, A. STIM1, PKC-δ and RasGRP set a threshold for proapoptotic Erk signaling during B cell development. Nat. Immunol. 2011, 12, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.; Savage, K.J.; Kutok, J.L.; Feuerhake, F.; Kurtin, P.; Mihm, M.; Wu, B.; Pasqualucci, L.; Neuberg, D.; Aguiar, R.C.T.; et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005, 105, 1851–1861. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.A.; Lee, M.; Hu, Y.; Andreas, J.; Patel, S.J.; Zhang, S.; Chines, P.; Elkahloun, A.; Chandrasekharappa, S.; Gutkind, J.S.; et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS Genet. 2014, 10, e1004809. [Google Scholar] [CrossRef]
- Umit, E.G.; Baysal, M.; Durmus, Y.; Demir, A.M. CD11c expression in chronic lymphocytic leukemia revisited, related with complications and survival. Int. J. Lab. Hematol. 2017, 39, 552–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, B.; Hu, W.-W.; Chen, L.-J.; Wu, C.-P.; Lu, B.-F.; Shen, Y.-P.; Jiang, J.-T. High expression of CD11c indicates favorable prognosis in patients with gastric cancer. World J. Gastroenterol. 2015, 21, 9403–9412. [Google Scholar] [CrossRef] [PubMed]
- Ohl, K.; Schippers, A.; Tenbrock, K. CD11c-Specific Deletion Reveals CREB as a Critical Regulator of DC Function during the Germinal Center Response. J. Immunol. Res. 2018, 2018, 8947230. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, N.; Mueller, M.; Mougiakakos, D.; Ihorst, G.; Marks, R.; Schmitt-Graeff, A.; Veelken, H. Analysis of dendritic cell subpopulations in follicular lymphoma with respect to the tumor immune microenvironment. Leuk. Lymphoma 2016, 57, 2150–2160. [Google Scholar] [CrossRef]
- Perez-Andres, M.; Paiva, B.; Nieto, W.G.; Caraux, A.; Schmitz, A.; Almeida, J.; Vogt, R.F.; Marti, G.E.; Rawstron, A.C.; Van Zelm, M.C.; et al. Human peripheral blood B-cell compartments: A crossroad in B-cell traffic. Cytom. Part B Clin. Cytom. 2010, 78, S47–S60. [Google Scholar] [CrossRef] [PubMed]
- Perbellini, O.; Falisi, E.; Giaretta, I.; Boscaro, E.; Novella, E.; Facco, M.; Fortuna, S.; Finotto, S.; Amati, E.; Maniscalco, F.; et al. Clinical significance of LAIR1 (CD305) as assessed by flow cytometry in a prospective series of patients with chronic lymphocytic leukemia. Haematologica 2014, 99, 881–887. [Google Scholar] [CrossRef] [Green Version]
- Menendez, P.; Vargas, A.; Bueno, C.; Barrena, S.; Almeida, J.; De Santiago, M.; López, A.; Roa, S.; San Miguel, J.F.; Orfao, A. Quantitative analysis of bcl-2 expression in normal and leukemic human B-cell differentiation. Leukemia 2004, 18, 491–498. [Google Scholar] [CrossRef]
- Kitano, M.; Moriyama, S.; Ando, Y.; Hikida, M.; Mori, Y.; Kurosaki, T.; Okada, T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity 2011, 34, 961–972. [Google Scholar] [CrossRef]
- Kurosu, T.; Fukuda, T.; Miki, T.; Miura, O. BCL6 overexpression prevents increase in reactive oxygen species and inhibits apoptosis induced by chemotherapeutic reagents in B-cell lymphoma cells. Oncogene 2003, 22, 4459–4468. [Google Scholar] [CrossRef] [Green Version]
- Dupont, T.; Yang, S.N.; Patel, J.; Hatzi, K.; Malik, A.; Tam, W.; Martin, P.; Leonard, J.; Melnick, A.; Cerchietti, L. Selective targeting of BCL6 induces oncogene addiction switching to BCL2 in B-cell lymphoma. Oncotarget 2016, 7, 3520–3532. [Google Scholar] [CrossRef]






| A. | Overall Survival (n = 233) | |
| Prognostic Variable | HR | p-Value |
| Age (>60 years) | 2.20 | <0.0001 |
| GCB-ABC molecular subgroups | 2.75 | <0.0001 |
| IPI | 1.79 | <0.0001 |
| DNA repair score | 3.87 | <0.0001 |
| Risk Score | 2.41 | <0.0001 |
| B. | Overall Survival (n = 233) | |
| Prognostic Variable | HR | p-Value |
| Risk Score | 2.248 | <0.0001 |
| Age (>60 years) | 1.846 | 0.03 |
| Risk Score | 2.269 | <0.0001 |
| GCB-ABC molecular subgroups | 1.359 | NS |
| Risk Score | 2.113 | <0.0001 |
| IPI | 1.582 | <0.0001 |
| Risk Score | 2.263 | <0.0001 |
| DNA repair score | 2.909 | <0.0001 |
| C. | Overall Survival (n = 233) | |
| Prognostic Variable | HR | p-Value |
| Age (>60 years) | 1.80 | NS |
| GCB-ABC molecular subgroups | 2.23 | NS |
| IPI | 0.20 | NS |
| DNA repair score | 3.44 | <0.0001 |
| Risk Score | 1.61 | 0.007 |
| A. | Overall Survival (n = 233) | |
| Lymphoid Marker | HR | p-Value |
| BCL2 | 2.11 | 0.006 |
| BCL6 | 0.26 | <0.0001 |
| CD39 | 2.1 | 0.01 |
| CD103 | 1.82 | 0.03 |
| CD11c | 0.27 | <0.0001 |
| LAIR1 | 2.24 | 0.004 |
| CD10 | 0.37 | <0.0001 |
| CD5 | 0.59 | 0.04 |
| B. | Overall Survival (n = 233) | |
| Lymphoid Marker | HR | p-Value |
| BCL2 | 1.96 | 0.01 |
| BCL6 | 0.44 | 0.007 |
| CD39 | 1.22 | NS |
| CD103 | 1.30 | NS |
| CD11c | 0.33 | <0.0001 |
| LAIR1 | 2.82 | 0.002 |
| CD10 | 0.39 | NS |
| CD5 | 0.47 | 0.01 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devin, J.; Kassambara, A.; Bruyer, A.; Moreaux, J.; Bret, C. Phenotypic Characterization of Diffuse Large B-Cell Lymphoma Cells and Prognostic Impact. J. Clin. Med. 2019, 8, 1074. https://doi.org/10.3390/jcm8071074
Devin J, Kassambara A, Bruyer A, Moreaux J, Bret C. Phenotypic Characterization of Diffuse Large B-Cell Lymphoma Cells and Prognostic Impact. Journal of Clinical Medicine. 2019; 8(7):1074. https://doi.org/10.3390/jcm8071074
Chicago/Turabian StyleDevin, Julie, Alboukadel Kassambara, Angélique Bruyer, Jérôme Moreaux, and Caroline Bret. 2019. "Phenotypic Characterization of Diffuse Large B-Cell Lymphoma Cells and Prognostic Impact" Journal of Clinical Medicine 8, no. 7: 1074. https://doi.org/10.3390/jcm8071074
APA StyleDevin, J., Kassambara, A., Bruyer, A., Moreaux, J., & Bret, C. (2019). Phenotypic Characterization of Diffuse Large B-Cell Lymphoma Cells and Prognostic Impact. Journal of Clinical Medicine, 8(7), 1074. https://doi.org/10.3390/jcm8071074