The Skin Microbiota and Itch: Is There a Link?
Abstract
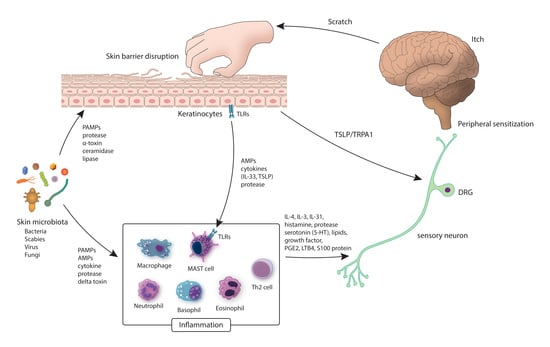
:1. Introduction
2. The Peripheral Mechanism Linking the Skin Microbiota and Itch
2.1. The Skin Microbiota, The Skin Barrier, and Itch
2.2. The Skin Microbiota, The Immune System, and Itch
2.3. The Skin Microbiota, The Sensory Nerve, and Itch
3. The Central Mechanism Linking the Skin Microbiota and Itch
3.1. Microbial Endocrinology
3.2. Stress, The Skin Microbiota, and Itch
3.3. The Skin Microbiota, The Amygdala, and Itch
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Glossary
| Term | Definition |
| 16S rRNA gene sequencing | Genomic analysis of 16S ribosomal RNA phylotypes from DNA that is extracted directly from bacterial communities in clinical or environmental samples, a process that circumvents culturing [29]. |
| Skin microbiota | Total of microbes in/on our skin [168]. |
| Microbiota | The group of microbes found in/on a specific environment or living host [169]. |
| Microbial diversity | Degree of variability of the microbiota. α-diversity describes within-sample variability, while β-diversity signifies variability between samples [169]. |
| Dysbiosis | Microbial imbalance or maladaptation [168]. |
| Prebiotics | Nutrients that stimulate beneficial skin microorganisms [170]. |
| Probiotics | Live microorganisms that have a favorable impact on host health when given in proper amounts [169]. |
| Antibiotics | Antibiotics block the growth of or destroy bacteria and other microbes [168]. |
References
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.J.; Chiu, I.M. Bacterial signaling to the nervous system through toxins and metabolites. J. Mol. Biol. 2017, 429, 587–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota-host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Gallo, R.L. Human skin is the largest epithelial surface for interaction with microbes. J. Investig. Dermatol. 2017, 137, 1213–1214. [Google Scholar] [CrossRef] [Green Version]
- Fyhrquist, N.; Salava, A.; Auvinen, P.; Lauerma, A. Skin biomes. Curr. Allergy Asthma Rep. 2016, 16, 40. [Google Scholar] [CrossRef] [Green Version]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [Green Version]
- Leyden, J.J.; McGinley, K.J.; Nordstrom, K.M.; Webster, G.F. Skin microflora. J. Investig. Dermatol. 1987, 88, 65s–72s. [Google Scholar] [CrossRef]
- Wang, W.M.; Jin, H.Z. Skin microbiome: An actor in the pathogenesis of psoriasis. Chin. Med. J. (Engl.) 2018, 131, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, B. Microbiome: The surface brigade. Nature 2012, 492, S60–S61. [Google Scholar] [CrossRef]
- Gontcharova, V.; Youn, E.; Sun, Y.; Wolcott, R.D.; Dowd, S.E. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol. J. 2010, 4, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.R.; Gomez, B.I.; McIntyre, M.K.; Dubick, M.A.; Christy, R.J.; Nicholson, S.E.; Burmeister, D.M. The cutaneous microbiome and wounds: New molecular targets to promote wound healing. Int. J. Mol. Sci. 2018, 19, 2699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, I.M. Infection, pain, and itch. Neurosci. Bull. 2018, 34, 109–119. [Google Scholar] [CrossRef]
- Kong, H.H.; Oh, J.; Deming, C.; Conlan, S.; Grice, E.A.; Beatson, M.A.; Nomicos, E.; Polley, E.C.; Komarow, H.D.; Murray, P.R.; et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012, 22, 850–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blicharz, L.; Usarek, P.; Mlynarczyk, G.; Skowronski, K.; Rudnicka, L.; Samochocki, Z. Is itch intensity in atopic dermatitis associated with skin colonization by Staphylococcus aureus? Indian J. Dermatol. 2020, 65, 17–21. [Google Scholar] [CrossRef]
- Allen, H.B.; Vaze, N.D.; Choi, C.; Hailu, T.; Tulbert, B.H.; Cusack, C.A.; Joshi, S.G. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 2014, 150, 260–265. [Google Scholar] [CrossRef] [Green Version]
- Azimi, E.; Xia, J.; Lerner, E.A. Peripheral mechanisms of itch. Curr. Probl. Dermatol. 2016, 50, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Baldwin, H.E.; Bhatia, N.D.; Friedman, A.; Eng, R.M.; Seite, S. The role of cutaneous microbiota harmony in maintaining a functional skin barrier. J. Drugs Dermatol. 2017, 16, 12–18. [Google Scholar] [CrossRef]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [Green Version]
- Nakatsuji, T.; Chiang, H.I.; Jiang, S.B.; Nagarajan, H.; Zengler, K.; Gallo, R.L. The microbiome extends to subepidermal compartments of normal skin. Nat. Commun. 2013, 4, 1431. [Google Scholar] [CrossRef] [Green Version]
- Proksch, E. pH in nature, humans and skin. J. Dermatol. 2018, 45, 1044–1052. [Google Scholar] [CrossRef] [PubMed]
- Boer, M.; Duchnik, E.; Maleszka, R.; Marchlewicz, M. Structural and biophysical characteristics of human skin in maintaining proper epidermal barrier function. Postepy Dermatol. Alergol. 2016, 33, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Niyonsaba, F.; Someya, A.; Hirata, M.; Ogawa, H.; Nagaoka, I. Evaluation of the effects of peptide antibiotics human beta-defensins-1/-2 and LL-37 on histamine release and prostaglandin D(2) production from mast cells. Eur. J. Immunol. 2001, 31, 1066–1075. [Google Scholar] [CrossRef]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Investig. Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, M. Staphylococcus epidermidis—The ‘accidental’ pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Shu, M.; Wang, Y.; Yu, J.; Kuo, S.; Coda, A.; Jiang, Y.; Gallo, R.L.; Huang, C.M. Fermentation of Propionibacterium acnes, a commensal bacterium in the human skin microbiome, as skin probiotics against methicillin-resistant Staphylococcus aureus. PLoS ONE 2013, 8, e55380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Korting, H.C.; Hubner, K.; Greiner, K.; Hamm, G.; Braun-Falco, O. Differences in the skin surface pH and bacterial microflora due to the long-term application of synthetic detergent preparations of pH 5.5 and pH 7.0. Results of a crossover trial in healthy volunteers. Acta Derm. Venereol. 1990, 70, 429–431. [Google Scholar] [PubMed]
- Ali, S.M.; Yosipovitch, G. Skin pH: From basic science to basic skin care. Acta Derm. Venereol. 2013, 93, 261–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Smeden, J.; Bouwstra, J.A. Stratum corneum lipids: Their role for the skin barrier function in healthy subjects and atopic dermatitis patients. Curr. Probl. Dermatol. 2016, 49, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.S. The role of microorganisms in atopic dermatitis. Clin. Exp. Immunol. 2006, 144, 1–9. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Okino, N.; Ito, M.; Imayama, S. Ceramidase activity in bacterial skin flora as a possible cause of ceramide deficiency in atopic dermatitis. Clin. Diagn. Lab. Immunol. 1999, 6, 101–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.E.; Kim, H.S. Microbiome of the skin and gut in atopic dermatitis (AD): Understanding the pathophysiology and finding novel management strategies. J. Clin. Med. 2019, 8, 444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swe, P.M.; Zakrzewski, M.; Kelly, A.; Krause, L.; Fischer, K. Scabies mites alter the skin microbiome and promote growth of opportunistic pathogens in a porcine model. PLoS Negl. Trop. Dis. 2014, 8, e2897. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Saunders, C.W.; Hu, P.; Grant, R.A.; Boekhout, T.; Kuramae, E.E.; Kronstad, J.W.; Deangelis, Y.M.; Reeder, N.L.; Johnstone, K.R.; et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc. Natl. Acad. Sci. USA 2007, 104, 18730–18735. [Google Scholar] [CrossRef] [Green Version]
- Mack, M.R.; Kim, B.S. The itch-scratch cycle: A neuroimmune perspective. Trends Immunol. 2018, 39, 980–991. [Google Scholar] [CrossRef]
- Potenzieri, C.; Undem, B.J. Basic mechanisms of itch. Clin. Exp. Allergy 2012, 42, 8–19. [Google Scholar] [CrossRef] [Green Version]
- Borgono, C.A.; Michael, I.P.; Komatsu, N.; Jayakumar, A.; Kapadia, R.; Clayman, G.L.; Sotiropoulou, G.; Diamandis, E.P. A potential role for multiple tissue kallikrein serine proteases in epidermal desquamation. J. Biol. Chem. 2007, 282, 3640–3652. [Google Scholar] [CrossRef] [Green Version]
- Yosipovitch, G.; Misery, L.; Proksch, E.; Metz, M.; Stander, S.; Schmelz, M. Skin barrier damage and itch: Review of mechanisms, topical management and future directions. Acta Derm. Venereol. 2019, 99, 1201–1209. [Google Scholar] [CrossRef] [Green Version]
- Komatsu, N.; Saijoh, K.; Kuk, C.; Liu, A.C.; Khan, S.; Shirasaki, F.; Takehara, K.; Diamandis, E.P. Human tissue kallikrein expression in the stratum corneum and serum of atopic dermatitis patients. Exp. Dermatol. 2007, 16, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Steinhoff, M.; Neisius, U.; Ikoma, A.; Fartasch, M.; Heyer, G.; Skov, P.S.; Luger, T.A.; Schmelz, M. Proteinase-activated receptor-2 mediates itch: A novel pathway for pruritus in human skin. J. Neurosci. 2003, 23, 6176–6180. [Google Scholar] [CrossRef]
- Stefansson, K.; Brattsand, M.; Roosterman, D.; Kempkes, C.; Bocheva, G.; Steinhoff, M.; Egelrud, T. Activation of proteinase-activated receptor-2 by human kallikrein-related peptidases. J. Investig. Dermatol. 2008, 128, 18–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, K.M.; Nattkemper, L.A.; Rosen, J.D.; Andersen, H.H.; Hsiang, J.; Romanelli, P.; Bernigaud, C.; Guillot, J.; Chosidow, O.; Yosipovitch, G. Non-histaminergic itch mediators elevated in the skin of a porcine model of scabies and of human scabies patients. J. Investig. Dermatol. 2019, 139, 971–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niyonsaba, F.; Ushio, H.; Hara, M.; Yokoi, H.; Tominaga, M.; Takamori, K.; Kajiwara, N.; Saito, H.; Nagaoka, I.; Ogawa, H.; et al. Antimicrobial peptides human beta-defensins and cathelicidin LL-37 induce the secretion of a pruritogenic cytokine IL-31 by human mast cells. J. Immunol. 2010, 184, 3526–3534. [Google Scholar] [CrossRef] [Green Version]
- Cevikbas, F.; Wang, X.; Akiyama, T.; Kempkes, C.; Savinko, T.; Antal, A.; Kukova, G.; Buhl, T.; Ikoma, A.; Buddenkotte, J.; et al. A sensory neuron-expressed IL-31 receptor mediates T helper cell-dependent itch: Involvement of TRPV1 and TRPA1. J. Allergy Clin. Immunol. 2014, 133, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized control of skin immunity by resident commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef] [Green Version]
- Chehoud, C.; Rafail, S.; Tyldsley, A.S.; Seykora, J.T.; Lambris, J.D.; Grice, E.A. Complement modulates the cutaneous microbiome and inflammatory milieu. Proc. Natl. Acad. Sci. USA 2013, 110, 15061–15066. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef] [Green Version]
- Stacy, A.; Belkaid, Y. Microbial guardians of skin health. Science 2019, 363, 227–228. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Di Nardo, A.; Nakatsuji, T.; Leichtle, A.; Yang, Y.; Cogen, A.L.; Wu, Z.R.; Hooper, L.V.; Schmidt, R.R.; Von Aulock, S.; et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat. Med. 2009, 15, 1377–1382. [Google Scholar] [CrossRef] [PubMed]
- Menzies, B.E.; Kenoyer, A. Signal transduction and nuclear responses in Staphylococcus aureus-induced expression of human beta-defensin 3 in skin keratinocytes. Infect. Immun. 2006, 74, 6847–6854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, M.; Tawaratsumida, K.; Kariya, H.; Aoyama, K.; Tamura, T.; Suda, Y. Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int. Immunol. 2006, 18, 355–362. [Google Scholar] [CrossRef]
- Bubeck Wardenburg, J.; Williams, W.A.; Missiakas, D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. USA 2006, 103, 13831–13836. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Ochoa, M.T.; Krutzik, S.R.; Takeuchi, O.; Uematsu, S.; Legaspi, A.J.; Brightbill, H.D.; Holland, D.; Cunliffe, W.J.; Akira, S.; et al. Activation of toll-like receptor 2 in acne triggers inflammatory cytokine responses. J. Immunol. 2002, 169, 1535–1541. [Google Scholar] [CrossRef] [Green Version]
- Linehan, J.L.; Harrison, O.J.; Han, S.J.; Byrd, A.L.; Vujkovic-Cvijin, I.; Villarino, A.V.; Sen, S.K.; Shaik, J.; Smelkinson, M.; Tamoutounour, S.; et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell 2018, 172, 784–796.e18. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Munoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.; McGavin, M.J.; Travers, J.B.; et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature 2013, 503, 397–401. [Google Scholar] [CrossRef] [Green Version]
- Williams, M.R.; Nakatsuji, T.; Gallo, R.L. Staphylococcus aureus: Master manipulator of the skin. Cell Host Microbe 2017, 22, 579–581. [Google Scholar] [CrossRef] [Green Version]
- Tebruegge, M.; Kuruvilla, M.; Margarson, I. Does the use of calamine or antihistamine provide symptomatic relief from pruritus in children with varicella zoster infection? Arch. Dis. Child. 2006, 91, 1035–1036. [Google Scholar] [CrossRef] [Green Version]
- McKenzie, R.C.; Sauder, D.N. Keratinocyte cytokines and growth factors. Functions in skin immunity and homeostasis. Dermatol. Clin. 1990, 8, 649–661. [Google Scholar] [CrossRef]
- Kollisch, G.; Kalali, B.N.; Voelcker, V.; Wallich, R.; Behrendt, H.; Ring, J.; Bauer, S.; Jakob, T.; Mempel, M.; Ollert, M. Various members of the Toll-like receptor family contribute to the innate immune response of human epidermal keratinocytes. Immunology 2005, 114, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Lebre, M.C.; Van der Aar, A.M.; Van Baarsen, L.; Van Capel, T.M.; Schuitemaker, J.H.; Kapsenberg, M.L.; De Jong, E.C. Human keratinocytes express functional Toll-like receptor 3, 4, 5, and 9. J. Investig. Dermatol. 2007, 127, 331–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olaru, F.; Jensen, L.E. Chemokine expression by human keratinocyte cell lines after activation of Toll-like receptors. Exp. Dermatol. 2010, 19, e314–e316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dainichi, T.; Kitoh, A.; Otsuka, A.; Nakajima, S.; Nomura, T.; Kaplan, D.H.; Kabashima, K. The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat. Immunol. 2018, 19, 1286–1298. [Google Scholar] [CrossRef]
- Hofmann, A.M.; Abraham, S.N. New roles for mast cells in modulating allergic reactions and immunity against pathogens. Curr. Opin. Immunol. 2009, 21, 679–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igawa, S.; Di Nardo, A. Skin microbiome and mast cells. Transl. Res. 2017, 184, 68–76. [Google Scholar] [CrossRef]
- Leon, A.; Rosen, J.D.; Hashimoto, T.; Fostini, A.C.; Paus, R.; Yosipovitch, G. Itching for an answer: A review of potential mechanisms of scalp itch in psoriasis. Exp. Dermatol. 2019, 28, 1397–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trier, A.M.; Mack, M.R.; Kim, B.S. The neuroimmune axis in skin sensation, inflammation, and immunity. J. Immunol. 2019, 202, 2829–2835. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.M.; Hill, R.Z.; Schwendinger-Schreck, J.; Deguine, J.; Brock, E.C.; Kucirek, N.; Rifi, Z.; Wei, J.; Gronert, K.; Brem, R.B.; et al. Neutrophils promote CXCR3-dependent itch in the development of atopic dermatitis. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Rosen, J.D.; Sanders, K.M.; Yosipovitch, G. Possible role of neutrophils in itch. Itch 2018, 3, e17. [Google Scholar] [CrossRef]
- Luo, J.; Feng, J.; Liu, S.; Walters, E.T.; Hu, H. Molecular and cellular mechanisms that initiate pain and itch. Cell. Mol. Life Sci. 2015, 72, 3201–3223. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, T.; Satoh, T.; Yokozeki, H. Pruritus in ordinary scabies: IL-31 from macrophages induced by overexpression of thymic stromal lymphopoietin and periostin. Allergy 2019, 74, 1727–1737. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Kursewicz, C.D.; Fayne, R.A.; Nanda, S.; Shah, S.M.; Nattkemper, L.; Yokozeki, H.; Yosipovitch, G. Mechanisms of itch in stasis dermatitis: Significant role of IL-31 from macrophages. J. Investig. Dermatol. 2020, 140, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Sonkoly, E.; Muller, A.; Lauerma, A.I.; Pivarcsi, A.; Soto, H.; Kemeny, L.; Alenius, H.; Dieu-Nosjean, M.C.; Meller, S.; Rieker, J.; et al. IL-31: A new link between T cells and pruritus in atopic skin inflammation. J. Allergy Clin. Immunol. 2006, 117, 411–417. [Google Scholar] [CrossRef]
- Baral, P.; Mills, K.; Pinho-Ribeiro, F.A.; Chiu, I.M. Pain and itch: Beneficial or harmful to antimicrobial defense? Cell Host Microbe 2016, 19, 755–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Gao, Y.J.; Ji, R.R. Emerging role of Toll-like receptors in the control of pain and itch. Neurosci. Bull. 2012, 28, 131–144. [Google Scholar] [CrossRef]
- Liu, T.; Xu, Z.Z.; Park, C.K.; Berta, T.; Ji, R.R. Toll-like receptor 7 mediates pruritus. Nat. Neurosci. 2010, 13, 1460–1462. [Google Scholar] [CrossRef]
- Liu, T.; Berta, T.; Xu, Z.Z.; Park, C.K.; Zhang, L.; Lu, N.; Liu, Q.; Liu, Y.; Gao, Y.J.; Liu, Y.C.; et al. TLR3 deficiency impairs spinal cord synaptic transmission, central sensitization, and pruritus in mice. J. Clin. Investig. 2012, 122, 2195–2207. [Google Scholar] [CrossRef]
- Diogenes, A.; Ferraz, C.C.; Akopian, A.N.; Henry, M.A.; Hargreaves, K.M. LPS sensitizes TRPV1 via activation of TLR4 in trigeminal sensory neurons. J. Dent. Res. 2011, 90, 759–764. [Google Scholar] [CrossRef]
- Calil, I.L.; Zarpelon, A.C.; Guerrero, A.T.; Alves-Filho, J.C.; Ferreira, S.H.; Cunha, F.Q.; Cunha, T.M.; Verri, W.A., Jr. Lipopolysaccharide induces inflammatory hyperalgesia triggering a TLR4/MyD88-dependent cytokine cascade in the mice paw. PLoS ONE 2014, 9, e90013. [Google Scholar] [CrossRef]
- Min, H.; Lee, H.; Lim, H.; Jang, Y.H.; Chung, S.J.; Lee, C.J.; Lee, S.J. TLR4 enhances histamine-mediated pruritus by potentiating TRPV1 activity. Mol. Brain 2014, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Meseguer, V.; Alpizar, Y.A.; Luis, E.; Tajada, S.; Denlinger, B.; Fajardo, O.; Manenschijn, J.A.; Fernandez-Pena, C.; Talavera, A.; Kichko, T.; et al. TRPA1 channels mediate acute neurogenic inflammation and pain produced by bacterial endotoxins. Nat. Commun. 2014, 5, 3125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Startek, J.B.; Talavera, K.; Voets, T.; Alpizar, Y.A. Differential interactions of bacterial lipopolysaccharides with lipid membranes: Implications for TRPA1-mediated chemosensation. Sci. Rep. 2018, 8, 12010. [Google Scholar] [CrossRef]
- Kashem, S.W.; Riedl, M.S.; Yao, C.; Honda, C.N.; Vulchanova, L.; Kaplan, D.H. Nociceptive sensory fibers drive interleukin-23 production from CD301b+ dermal dendritic cells and drive protective cutaneous immunity. Immunity 2015, 43, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Chiu, I.M.; Heesters, B.A.; Ghasemlou, N.; Von Hehn, C.A.; Zhao, F.; Tran, J.; Wainger, B.; Strominger, A.; Muralidharan, S.; Horswill, A.R.; et al. Bacteria activate sensory neurons that modulate pain and inflammation. Nature 2013, 501, 52–57. [Google Scholar] [CrossRef] [Green Version]
- Pinho-Ribeiro, F.A.; Baddal, B.; Haarsma, R.; O’Seaghdha, M.; Yang, N.J.; Blake, K.J.; Portley, M.; Verri, W.A.; Dale, J.B.; Wessels, M.R.; et al. Blocking neuronal signaling to immune cells treats streptococcal invasive infection. Cell 2018, 173, 1083–1097.e22. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, T.; Yosipovitch, G. Itching as a systemic disease. J. Allergy Clin. Immunol. 2019, 144, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, J.S.; Fagan, A.; Sikaroodi, M.; Kakiyama, G.; Takei, H.; Degefu, Y.; Pandak, W.M.; Hylemon, P.B.; Fuchs, M.; John, B.; et al. Alterations in skin microbiomes of patients with cirrhosis. Clin. Gastroenterol. Hepatol. 2019, 17, 2581–2591.e15. [Google Scholar] [CrossRef]
- Kremer, A.E.; Martens, J.J.; Kulik, W.; Rueff, F.; Kuiper, E.M.; Van Buuren, H.R.; Van Erpecum, K.J.; Kondrackiene, J.; Prieto, J.; Rust, C.; et al. Lysophosphatidic acid is a potential mediator of cholestatic pruritus. Gastroenterology 2010, 139, 1008–1018.e1. [Google Scholar] [CrossRef] [Green Version]
- Beuers, U.; Kremer, A.E.; Bolier, R.; Elferink, R.P. Pruritus in cholestasis: Facts and fiction. Hepatology 2014, 60, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Posadas, A.; Picazo-Juarez, G.; Llorente, I.; Jara-Oseguera, A.; Morales-Lazaro, S.; Escalante-Alcalde, D.; Islas, L.D.; Rosenbaum, T. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nat. Chem. Biol. 2011, 8, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Roosterman, D.; Goerge, T.; Schneider, S.W.; Bunnett, N.W.; Steinhoff, M. Neuronal control of skin function: The skin as a neuroimmunoendocrine organ. Physiol. Rev. 2006, 86, 1309–1379. [Google Scholar] [CrossRef] [PubMed]
- Pereira, U.; Boulais, N.; Lebonvallet, N.; Lefeuvre, L.; Gougerot, A.; Misery, L. Development of an in vitro coculture of primary sensitive pig neurons and keratinocytes for the study of cutaneous neurogenic inflammation. Exp. Dermatol. 2010, 19, 931–935. [Google Scholar] [CrossRef] [PubMed]
- Mijouin, L.; Hillion, M.; Ramdani, Y.; Jaouen, T.; Duclairoir-Poc, C.; Follet-Gueye, M.L.; Lati, E.; Yvergnaux, F.; Driouich, A.; Lefeuvre, L.; et al. Effects of a skin neuropeptide (substance p) on cutaneous microflora. PLoS ONE 2013, 8, e78773. [Google Scholar] [CrossRef] [PubMed]
- N’Diaye, A.; Gannesen, A.; Borrel, V.; Maillot, O.; Enault, J.; Racine, P.J.; Plakunov, V.; Chevalier, S.; Lesouhaitier, O.; Feuilloley, M.G. Substance P and calcitonin gene-related peptide: Key regulators of cutaneous microbiota homeostasis. Front. Endocrinol. (Lausanne) 2017, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- N’Diaye, A.; Mijouin, L.; Hillion, M.; Diaz, S.; Konto-Ghiorghi, Y.; Percoco, G.; Chevalier, S.; Lefeuvre, L.; Harmer, N.J.; Lesouhaitier, O.; et al. Effect of substance P in Staphylococcus aureus and Staphylococcus epidermidis virulence: Implication for skin homeostasis. Front. Microbiol. 2016, 7, 506. [Google Scholar] [CrossRef]
- Raap, U.; Stander, S.; Metz, M. Pathophysiology of itch and new treatments. Curr. Opin. Allergy Clin. Immunol. 2011, 11, 420–427. [Google Scholar] [CrossRef]
- Lenard, J. Mammalian hormones in microbial cells. Trends Biochem. Sci. 1992, 17, 147–150. [Google Scholar] [CrossRef]
- Kawashima, K.; Misawa, H.; Moriwaki, Y.; Fujii, Y.X.; Fujii, T.; Horiuchi, Y.; Yamada, T.; Imanaka, T.; Kamekura, M. Ubiquitous expression of acetylcholine and its biological functions in life forms without nervous systems. Life Sci. 2007, 80, 2206–2209. [Google Scholar] [CrossRef]
- Stephenson, M.; Rowatt, E. The production of acetylcholine by a strain of Lactobacillus plantarum. J. Gen. Microbiol. 1947, 1, 279–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masson, F.; Talon, R.; Montel, M.C. Histamine and tyramine production by bacteria from meat products. Int. J. Food Microbiol. 1996, 32, 199–207. [Google Scholar] [CrossRef]
- Thomas, C.M.; Hong, T.; Van Pijkeren, J.P.; Hemarajata, P.; Trinh, D.V.; Hu, W.; Britton, R.A.; Kalkum, M.; Versalovic, J. Histamine derived from probiotic Lactobacillus reuteri suppresses TNF via modulation of PKA and ERK signaling. PLoS ONE 2012, 7, e31951. [Google Scholar] [CrossRef] [Green Version]
- Hurley, R.; Leask, B.G.; Ruthven, C.R.; Sandler, M.; Southgate, J. Investigation of 5-hydroxytryptamine production by Candida albicans in vitro and in vivo. Microbios 1971, 4, 133–143. [Google Scholar] [PubMed]
- Asano, Y.; Hiramoto, T.; Nishino, R.; Aiba, Y.; Kimura, T.; Yoshihara, K.; Koga, Y.; Sudo, N. Critical role of gut microbiota in the production of biologically active, free catecholamines in the gut lumen of mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1288–G1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsavkelova, E.A.; Botvinko, I.V.; Kudrin, V.S.; Oleskin, A.V. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl. Biochem. 2000, 372, 115–117. [Google Scholar] [PubMed]
- Raasch, W.; Regunathan, S.; Li, G.; Reis, D.J. Agmatine, the bacterial amine, is widely distributed in mammalian tissues. Life Sci. 1995, 56, 2319–2330. [Google Scholar] [CrossRef]
- Arena, M.E.; Manca de Nadra, M.C. Biogenic amine production by Lactobacillus. J. Appl. Microbiol. 2001, 90, 158–162. [Google Scholar] [CrossRef] [Green Version]
- Leroith, D.; Liotta, A.S.; Roth, J.; Shiloach, J.; Lewis, M.E.; Pert, C.B.; Krieger, D.T. Corticotropin and beta-endorphin-like materials are native to unicellular organisms. Proc. Natl. Acad. Sci. USA 1982, 79, 2086–2090. [Google Scholar] [CrossRef] [Green Version]
- LeRoith, D.; Pickens, W.; Vinik, A.I.; Shiloach, J. Bacillus subtilis contains multiple forms of somatostatin-like material. Biochem. Biophys. Res. Commun. 1985, 127, 713–719. [Google Scholar] [CrossRef]
- Schar, G.; Stover, E.P.; Clemons, K.V.; Feldman, D.; Stevens, D.A. Progesterone binding and inhibition of growth in Trichophyton mentagrophytes. Infect. Immun. 1986, 52, 763–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef]
- SM, O.M.; Dinan, T.G.; Cryan, J.F. The gut microbiota as a key regulator of visceral pain. Pain 2017, 158 (Suppl. 1), S19–S28. [Google Scholar] [CrossRef] [Green Version]
- Holzer, P.; Farzi, A. Neuropeptides and the microbiota-gut-brain axis. Adv. Exp. Med. Biol. 2014, 817, 195–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yosipovitch, G.; Ansari, N.; Goon, A.; Chan, Y.H.; Goh, C.L. Clinical characteristics of pruritus in chronic idiopathic urticaria. Br. J. Dermatol. 2002, 147, 32–36. [Google Scholar] [CrossRef] [PubMed]
- Yosipovitch, G.; Goon, A.; Wee, J.; Chan, Y.H.; Goh, C.L. The prevalence and clinical characteristics of pruritus among patients with extensive psoriasis. Br. J. Dermatol. 2000, 143, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Yosipovitch, G.; Goon, A.T.; Wee, J.; Chan, Y.H.; Zucker, I.; Goh, C.L. Itch characteristics in Chinese patients with atopic dermatitis using a new questionnaire for the assessment of pruritus. Int. J. Dermatol. 2002, 41, 212–216. [Google Scholar] [CrossRef]
- Golpanian, R.S.; Kim, H.S.; Yosipovitch, G. Effects of stress on itch. Clin. Ther. 2020. [Google Scholar] [CrossRef]
- Yosipovitch, G.; Mochizuki, H. Neuroimaging of itch as a tool of assessment of chronic itch and its management. Handb. Exp. Pharmacol. 2015, 226, 57–70. [Google Scholar] [CrossRef]
- Galley, J.D.; Nelson, M.C.; Yu, Z.; Dowd, S.E.; Walter, J.; Kumar, P.S.; Lyte, M.; Bailey, M.T. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 2014, 14, 189. [Google Scholar] [CrossRef] [Green Version]
- Bailey, M.T. Influence of stressor-induced nervous system activation on the intestinal microbiota and the importance for immunomodulation. Adv. Exp. Med. Biol. 2014, 817, 255–276. [Google Scholar] [CrossRef] [PubMed]
- Slominski, A. A nervous breakdown in the skin: Stress and the epidermal barrier. J. Clin. Investig. 2007, 117, 3166–3169. [Google Scholar] [CrossRef] [Green Version]
- Aberg, K.M.; Radek, K.A.; Choi, E.H.; Kim, D.K.; Demerjian, M.; Hupe, M.; Kerbleski, J.; Gallo, R.L.; Ganz, T.; Mauro, T.; et al. Psychological stress downregulates epidermal antimicrobial peptide expression and increases severity of cutaneous infections in mice. J. Clin. Investig. 2007, 117, 3339–3349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radek, K.A. Antimicrobial anxiety: The impact of stress on antimicrobial immunity. J. Leukoc. Biol. 2010, 88, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Yosipovitch, G. An aberrant parasympathetic response: A new perspective linking chronic stress and itch. Exp. Dermatol. 2013, 22, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.W.; Papoiu, A.D.; Russoniello, C.V.; Wang, H.; Patel, T.S.; Chan, Y.H.; Yosipovitch, G. Effect of itch, scratching and mental stress on autonomic nervous system function in atopic dermatitis. Acta Derm. Venereol. 2010, 90, 354–361. [Google Scholar] [CrossRef]
- Curtis, B.J.; Radek, K.A. Cholinergic regulation of keratinocyte innate immunity and permeability barrier integrity: New perspectives in epidermal immunity and disease. J. Investig. Dermatol. 2012, 132, 28–42. [Google Scholar] [CrossRef] [Green Version]
- Radek, K.A.; Elias, P.M.; Taupenot, L.; Mahata, S.K.; O’Connor, D.T.; Gallo, R.L. Neuroendocrine nicotinic receptor activation increases susceptibility to bacterial infections by suppressing antimicrobial peptide production. Cell Host Microbe 2010, 7, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Curtis, B.J.; Plichta, J.K.; Blatt, H.; Droho, S.; Griffin, T.M.; Radek, K.A. Nicotinic acetylcholine receptor stimulation impairs epidermal permeability barrier function and recovery and modulates cornified envelope proteins. Life Sci. 2012, 91, 1070–1076. [Google Scholar] [CrossRef] [Green Version]
- Lyte, M.; Freestone, P.P.; Neal, C.P.; Olson, B.A.; Haigh, R.D.; Bayston, R.; Williams, P.H. Stimulation of Staphylococcus epidermidis growth and biofilm formation by catecholamine inotropes. Lancet 2003, 361, 130–135. [Google Scholar] [CrossRef]
- Freestone, P.P.; Haigh, R.D.; Williams, P.H.; Lyte, M. Stimulation of bacterial growth by heat-stable, norepinephrine-induced autoinducers. FEMS Microbiol. Lett. 1999, 172, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Neal, C.P.; Freestone, P.P.; Maggs, A.F.; Haigh, R.D.; Williams, P.H.; Lyte, M. Catecholamine inotropes as growth factors for Staphylococcus epidermidis and other coagulase-negative staphylococci. FEMS Microbiol. Lett. 2001, 194, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Borrel, V.; Thomas, P.; Catovic, C.; Racine, P.J.; Konto-Ghiorghi, Y.; Lefeuvre, L.; Duclairoir-Poc, C.; Zouboulis, C.C.; Feuilloley, M.G.J. Acne and stress: Impact of catecholamines on Cutibacterium acnes. Front. Med. (Lausanne) 2019, 6, 155. [Google Scholar] [CrossRef] [PubMed]
- Clarke, S.R.; Mohamed, R.; Bian, L.; Routh, A.F.; Kokai-Kun, J.F.; Mond, J.J.; Tarkowski, A.; Foster, S.J. The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 2007, 1, 199–212. [Google Scholar] [CrossRef] [Green Version]
- Freestone, P.P.; Sandrini, S.M.; Haigh, R.D.; Lyte, M. Microbial endocrinology: How stress influences susceptibility to infection. Trends Microbiol. 2008, 16, 55–64. [Google Scholar] [CrossRef]
- Pastar, I.; Nusbaum, A.G.; Gil, J.; Patel, S.B.; Chen, J.; Valdes, J.; Stojadinovic, O.; Plano, L.R.; Tomic-Canic, M.; Davis, S.C. Interactions of methicillin resistant Staphylococcus aureus USA300 and Pseudomonas aeruginosa in polymicrobial wound infection. PLoS ONE 2013, 8, e56846. [Google Scholar] [CrossRef]
- Choi, E.H.; Demerjian, M.; Crumrine, D.; Brown, B.E.; Mauro, T.; Elias, P.M.; Feingold, K.R. Glucocorticoid blockade reverses psychological stress-induced abnormalities in epidermal structure and function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 291, R1657–R1662. [Google Scholar] [CrossRef] [Green Version]
- Sandrini, S.M.; Shergill, R.; Woodward, J.; Muralikuttan, R.; Haigh, R.D.; Lyte, M.; Freestone, P.P. Elucidation of the mechanism by which catecholamine stress hormones liberate iron from the innate immune defense proteins transferrin and lactoferrin. J. Bacteriol. 2010, 192, 587–594. [Google Scholar] [CrossRef] [Green Version]
- Shibata, M.; Katsuyama, M.; Onodera, T.; Ehama, R.; Hosoi, J.; Tagami, H. Glucocorticoids enhance Toll-like receptor 2 expression in human keratinocytes stimulated with Propionibacterium acnes or proinflammatory cytokines. J. Investig. Dermatol. 2009, 129, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Seth, A.K.; Geringer, M.R.; Nguyen, K.T.; Agnew, S.P.; Dumanian, Z.; Galiano, R.D.; Leung, K.P.; Mustoe, T.A.; Hong, S.J. Bacteriophage therapy for Staphylococcus aureus biofilm-infected wounds: A new approach to chronic wound care. Plast. Reconstr. Surg. 2013, 131, 225–234. [Google Scholar] [CrossRef]
- Rojas, I.G.; Padgett, D.A.; Sheridan, J.F.; Marucha, P.T. Stress-induced susceptibility to bacterial infection during cutaneous wound healing. Brain Behav. Immun. 2002, 16, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Sonnex, C. Influence of ovarian hormones on urogenital infection. Sex. Transm. Infect. 1998, 74, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veinante, P.; Yalcin, I.; Barrot, M. The amygdala between sensation and affect: A role in pain. J. Mol. Psychiatry 2013, 1, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neugebauer, V.; Li, W. Differential sensitization of amygdala neurons to afferent inputs in a model of arthritic pain. J. Neurophysiol. 2003, 89, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, V.; Li, W.; Bird, G.C.; Han, J.S. The amygdala and persistent pain. Neuroscientist 2004, 10, 221–234. [Google Scholar] [CrossRef]
- Sanders, K.M.; Akiyama, T. The vicious cycle of itch and anxiety. Neurosci. Biobehav Rev. 2018, 87, 17–26. [Google Scholar] [CrossRef]
- Mu, D.; Deng, J.; Liu, K.F.; Wu, Z.Y.; Shi, Y.F.; Guo, W.M.; Mao, Q.Q.; Liu, X.J.; Li, H.; Sun, Y.G. A central neural circuit for itch sensation. Science 2017, 357, 695–699. [Google Scholar] [CrossRef] [Green Version]
- Jeong, K.Y.; Kang, J.H. Investigation of the pruritus-induced functional activity in the rat brain using manganese-enhanced MRI. J. Magn. Reson. Imaging 2015, 42, 709–716. [Google Scholar] [CrossRef]
- Davidson, S.; Zhang, X.; Khasabov, S.G.; Simone, D.A.; Giesler, G.J., Jr. Relief of itch by scratching: State-dependent inhibition of primate spinothalamic tract neurons. Nat. Neurosci. 2009, 12, 544–546. [Google Scholar] [CrossRef]
- Roozendaal, B.; McEwen, B.S.; Chattarji, S. Stress, memory and the amygdala. Nat. Rev. Neurosci. 2009, 10, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Pavlenko, D.; Akiyama, T. Why does stress aggravate itch? A possible role of the amygdala. Exp. Dermatol. 2019, 28, 1439–1441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mochizuki, H.; Hernandez, L.E.; Yosipovitch, G. What does brain imaging tell us about itch? Itch 2019, 4, e23. [Google Scholar] [CrossRef]
- Cowan, C.S.M.; Hoban, A.E.; Ventura-Silva, A.P.; Dinan, T.G.; Clarke, G.; Cryan, J.F. Gutsy moves: The amygdala as a critical node in microbiota to brain signaling. Bioessays 2018, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stilling, R.M.; Ryan, F.J.; Hoban, A.E.; Shanahan, F.; Clarke, G.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Microbes & neurodevelopment—Absence of microbiota during early life increases activity-related transcriptional pathways in the amygdala. Brain Behav. Immun. 2015, 50, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Hoban, A.E.; Stilling, R.M.; Moloney, G.M.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F.; Clarke, G. Microbial regulation of microRNA expression in the amygdala and prefrontal cortex. Microbiome 2017, 5, 102. [Google Scholar] [CrossRef] [Green Version]
- Luczynski, P.; Tramullas, M.; Viola, M.; Shanahan, F.; Clarke, G.; O’Mahony, S.; Dinan, T.G.; Cryan, J.F. Microbiota regulates visceral pain in the mouse. Elife 2017, 6. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Onaolapo, O.J.; Onaolapo, A.Y.; Olowe, A.O. The neurobehavioral implications of the brain and microbiota interaction. Front. Biosci. (Landmark Ed.) 2020, 25, 363–397. [Google Scholar] [CrossRef]
- O’Neill, C.A.; Monteleone, G.; McLaughlin, J.T.; Paus, R. The gut-skin axis in health and disease: A paradigm with therapeutic implications. Bioessays 2016, 38, 1167–1176. [Google Scholar] [CrossRef] [PubMed]
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The gut microbiome as a major regulator of the gut-skin axis. Front. Microbiol. 2018, 9, 1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arck, P.; Handjiski, B.; Hagen, E.; Pincus, M.; Bruenahl, C.; Bienenstock, J.; Paus, R. Is there a ‘gut-brain-skin axis’? Exp. Dermatol. 2010, 19, 401–405. [Google Scholar] [CrossRef]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential role of the microbiome in acne: A comprehensive review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef] [Green Version]
- Sanders, K.M.; Nattkemper, L.A.; Yosipovitch, G. The gut-itch connection. Exp. Dermatol. 2016, 25, 344–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro, J.; Harrington, A.M.; Lieu, T.; Garcia-Caraballo, S.; Maddern, J.; Schober, G.; O’Donnell, T.; Grundy, L.; Lumsden, A.L.; Miller, P.; et al. Activation of pruritogenic TGR5, MrgprA3, and MrgprC11 on colon-innervating afferents induces visceral hypersensitivity. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Egert, M.; Simmering, R.; Riedel, C.U. The association of the skin microbiota with health, immunity, and disease. Clin. Pharmacol. Ther. 2017, 102, 62–69. [Google Scholar] [CrossRef]
- Dreno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef]
- Bastiaanssen, T.F.S.; Cowan, C.S.M.; Claesson, M.J.; Dinan, T.G.; Cryan, J.F. Making sense of … the microbiome in psychiatry. Int. J. Neuropsychopharmacol. 2019, 22, 37–52. [Google Scholar] [CrossRef]
- Davani-Davari, D.; Negahdaripour, M.; Karimzadeh, I.; Seifan, M.; Mohkam, M.; Masoumi, S.J.; Berenjian, A.; Ghasemi, Y. Prebiotics: Definition, types, sources, mechanisms, and clinical applications. Foods 2019, 8, 92. [Google Scholar] [CrossRef] [Green Version]




| Bacteria | Interactions with TLRs |
|---|---|
| S. epidermidis | Adjusts TLR3-dependent inflammation by introducing a TLR2-mediated crosstalk to subdue inflammation [52]. Elicits keratinocytes to display AMPs through a TLR2-dependent mechanism [50]. |
| S. aureus | Induction of hBD3 gene expression is TLR2-dependent [53]. Lipoteichoic acid and bacterial lipoproteins act as TLR2/2 or TLR2/6 agonists [54,55]. |
| P. acnes | Colonizes sebaceous glands and stimulates KCs to release inflammatory cytokines via TLR2 activation [56]. |
| Bacteria | Effects of Stress Mediators |
|---|---|
| Staphylococcus epidermidis | Glucocorticoids decrease the effects of super antigen activated T cells and inhibit staphylococcal exotoxin-induced T cell proliferation, cytokine secretion [137]. Catecholamines induce biofilm growth [130]. |
| Propionibacterium acnes | Cortisol and steroids significantly exacerbate inflammation associated with P. acnes via TLR2 stimulation [138,139]. |
| Pseudomonas aeruginosa | Norepinephrine increases expression of the attachment factor PA-1 of P. aeruginosa and increase biofilm formation [135,138]. |
| Staphylococcus aureus | Acetylcholine augments susceptibility to infection by S. aureus [124]. Norepinephrine increases S. aureus’ ability to remove iron from host and therefore facilitates the bacteria to form biofilms [138,140]. |
| Group A Streptococcus | Cortisol alters vulnerability to Group A Streptococcus pyogenes skin infection [141]. Acetylcholine augments susceptibility to infection by Group A Streptococcus [124]. Catecholamines raise Staphylococcal growth by 5-log orders [130,131,132]. Catecholamines enhance Group A Streptococcus growth likely by increasing iron availability [138,142]. |
| Candida | Estrogen enhances Candida infectivity, switching yeast form to an invasive hyphae [143]. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.S.; Yosipovitch, G. The Skin Microbiota and Itch: Is There a Link? J. Clin. Med. 2020, 9, 1190. https://doi.org/10.3390/jcm9041190
Kim HS, Yosipovitch G. The Skin Microbiota and Itch: Is There a Link? Journal of Clinical Medicine. 2020; 9(4):1190. https://doi.org/10.3390/jcm9041190
Chicago/Turabian StyleKim, Hei Sung, and Gil Yosipovitch. 2020. "The Skin Microbiota and Itch: Is There a Link?" Journal of Clinical Medicine 9, no. 4: 1190. https://doi.org/10.3390/jcm9041190
APA StyleKim, H. S., & Yosipovitch, G. (2020). The Skin Microbiota and Itch: Is There a Link? Journal of Clinical Medicine, 9(4), 1190. https://doi.org/10.3390/jcm9041190