Roles of Nicotinamide Adenine Dinucleotide (NAD+) in Biological Systems
Abstract
:1. Introduction
1.1. Nicotinamide Adenine Dinucleotide (NAD+)
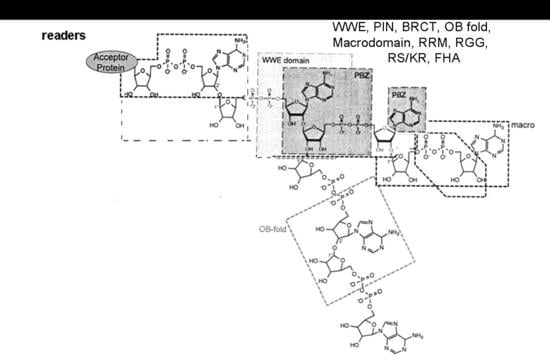
1.2. NAD+ Dependent ADP Ribosylation and Deacetylation Reactions
1.3. Bacterial Toxins
1.4. ADPRTs Modifying Nucleic Acids
1.5. Sirtuins
1.6. Viruses and Virulence
1.7. NAD+ Recycling, Localized NAD+ Consumption, and Effects of Defective NAD+ Metabolism
1.8. Non Enzymatic ADP-Ribose Attachment
1.9. Ap4A and Oligoadenylates
1.10. Nicotinic Acid Adenine Dinucleotide Phosphate (NAADP) and Cyclic ADP Ribose (cADPr)
2. Plants: Role of NAD+ and PARP Domain Enzymes in Immune Response
2.1. Extracellular NAD+ as DAMP Signal in the Apoplast: Plant NAD+ Receptors on Cell Membrane
2.2. Bacterial Effectors, PARP Domain Proteins, NAD+ and Plant Immunity
2.3. Hypothesis for the Role of NAD+: Convergence of Signaling Pathways in Plant Systems
3. Conclusions
Author Contributions
Conflicts of Interest
References
- Poltronieri, P.; Miwa, M. Overview on ADP ribosylation and PARP superfamily of proteins. Curr. Protein Pept. Sci. 2016, 17, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Poltronieri, P. ADP ribosylation reactions in animals, plants and bacteria. Challenges 2017, 8, 14. [Google Scholar] [CrossRef]
- Bürkle, A.; Virág, L. Poly(ADP-ribose): PARadigms and PARadoxes. Mol. Asp. Med. 2013, 34, 1046–1065. [Google Scholar] [CrossRef] [PubMed]
- Miwa, M.; Ida, C.; Yamashita, S.; Tanaka, M.; Fujisawa, J. Poly(ADP-ribose): Structure, physicochemical properties and quantification in vivo, with special reference to Poly(ADP-ribose) binding protein modules. Curr. Protein Pept. Sci. 2016, 17, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Verheugd, P.; Bütepage, M.; Eckei, L.; Lüscher, B. Players in ADP-ribosylation: Readers and Erasers. Curr. Protein Pept. Sci. 2016, 17, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Fouquerel, E.; Sobol, R.W. ARTD1 (PARP1) activation and NAD(+) in DNA repair and cell death. DNA Repair 2014, 23, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Bock, F.J.; Chang, P. Stress response. PARP1 911. Nat. Chem. Biol. 2015, 11, 179–180. [Google Scholar] [CrossRef] [PubMed]
- Faraone-Mennella, M.R. A new facet of ADP-ribosylation reactions: SIRTs and PARPs interplay. Front. Biosci. 2015, 20, 458–473. [Google Scholar] [CrossRef]
- Aravind, L.; Zhang, D.; de Souza, R.F.; Anand, S.; Iyer, L.M. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 2015, 384, 3–32. [Google Scholar] [PubMed]
- Timinszky, G.; Till, S.; Hassa, P.O.; Hothorn, M.; Kustatscher, G.; Nijmeijer, B.; Colombelli, J.; Altmeyer, M.; Stelzer, E.H.; Scheffzek, K.; et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009, 16, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.S.; Jividen, K.; Spencer, A.; Dworak, N.; Ni, L.; Oostdyk, L.T.; Chatterjee, M.; Kusmider, B.; Reon, B.; Parlak, M.; Gorbunova, V.; Abbas, T.; Jeffery, E.; Sherman, N.E.; Paschal, B.M. Ubiquitin modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 2017, 66, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Schmoll, M.; Dattenbock, C.; Carreras-Villasenor, N.; Mendoza-Mendoza, A.; Tisch, D.; Alemán, M.I.; Baker, S.E.; Brown, C.; Cervantes-Badillo, M.G.; Cetz-Chel, J.; et al. The genomes of three uneven siblings: Footprints of the lifestyles of three Trichoderma species. Microbiol. Mol. Biol. Rev. 2016, 80, 205–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rissel, D.; Heym, P.P.; Peiter, E. A yeast growth assay to characterize plant poly(ADP-ribose) polymerase (PARP) proteins and inhibitors. Anal. Biochem. 2017, 527, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Tao, Z.; Gao, P.; Liu, H.W. Studies of the expression of human poly(ADP-ribose) polymerase-1 in Saccharomyces cerevisiae and identification of PARP-1 substrates by yeast proteome microarray screening. Biochemistry 2009, 48, 11745–11754. [Google Scholar] [CrossRef] [PubMed]
- La Ferla, M.; Mercatanti, A.; Rocchi, G.; Lodovichi, S.; Cervelli, T.; Pignata, L.; Caligo, M.A.; Galli, A. Expression of human poly (ADP-ribose) polymerase 1 in Saccharomyces cerevisiae: Effect on survival, homologous recombination and identification of genes involved in intracellular localization. Mutat. Res. 2015, 774, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Lu, D.; Kabbage, M.; Wei, H.L.; Swingle, B.; Records, A.R.; Dickman, M.; He, P.; Shan, L. Bacterial effector HopF2 suppresses Arabidopsis innate immunity at the plasma membrane. Mol. Plant Microbe Interact. 2011, 24, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Durán, R.; Bourdais, G.; He, S.Y.; Robatzek, S. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytol. 2014, 202, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Jankevicius, G.; Ariza, A.; Ahel, M.; Ahel, I. The Toxin-Antitoxin system DarTG catalyzes reversible ADP-Ribosylation of DNA. Mol. Cell 2016, 64, 1109–1116. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, I.; Pellegrino Coppola, D.; Strippoli, M.; Ronzulli, R.; Vai, M. Nicotinamide supplementation phenocopies SIR2 inactivation by modulating carbon metabolism and respiration during yeast chronological aging. Mech. Ageing Dev. 2017, 161, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Mittal, N.; Muthuswami, R.; Madhubala, R. The mitochondrial SIR2 related protein 2 (SIR2RP2) impacts Leishmania donovani growth and infectivity. PLoS Negl. Trop. Dis. 2017, 11, e0005590. [Google Scholar] [CrossRef] [PubMed]
- Rack, J.G.M.; Morra, R.; Barkauskaite, E.; Kraehenbuehl, R.; Ariza, A.; Qu, Y.; Ortmayer, M.; Leidecker, O.; Cameron, D.R.; Matic, I.; et al. Identification of a class of protein ADP-ribosylating sirtuins in microbial pathogens. Mol. Cell 2015, 59, 309–320. [Google Scholar] [CrossRef] [PubMed]
- McPherson, R.L.; Abraham, R.; Sreekumar, E.; Ong, S.E.; Cheng, S.J.; Baxter, V.K.; Kistemaker, H.A.; Filippov, D.V.; Griffin, D.E.; Leung, A.K. ADP-ribosylhydrolase activity of Chikungunya virus macrodomain is critical for virus replication and virulence. Proc. Natl. Acad. Sci. USA 2017, 114, 1666–1671. [Google Scholar] [CrossRef] [PubMed]
- Steffen, J.D.; Pascal, J.M. New players to the field of ADP-ribosylation make the final cut. EMBO J. 2013, 32, 1225–1237. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, C.; Song, Y.; Shao, C.; Zhang, X.; Zang, J. Structural insights into the mechanism of Escherichia coli YmdB: A 2’-O-acetyl-ADP-ribose deacetylase. J. Struct. Biol. 2015, 192, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Haikarainen, T.; Lehtiö, L. Proximal ADP-ribose hydrolysis in Trypanosomatids is catalyzed by a macrodomain. Sci. Rep. 2016, 6, 24213. [Google Scholar] [CrossRef] [PubMed]
- Hwang, E.S.; Song, S.B. Nicotinamide is an inhibitor of SIRT1 in vitro, but can be a stimulator in cells. Cell Mol. Life Sci. 2017, 74, 3347–3362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Berrocal, J.G.; Frizzell, K.M.; Gamble, M.J.; DuMond, M.E.; Krishnakumar, R.; Yang, T.; Sauve, A.A.; Kraus, W.L. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J. Biol. Chem. 2009, 284, 20408–20417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Berrocal, J.G.; Yao, J.; DuMond, M.E.; Krishnakumar, R.; Ruhl, D.D.; Ryu, K.W.; Gamble, M.J.; Kraus, W.L. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J. Biol. Chem. 2012, 287, 12405–12416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Kraus, W.L. SIRT1-dependent regulation of chromatin and transcription: Linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim. Biophys. Acta 2010, 1804, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, M.; Masuo, S.; Fujita, T.; Doi, Y.; Kamimura, Y.; Takaya, N. Hydrolase controls cellular NAD, sirtuin, and secondary metabolites. Mol. Cell Biol. 2012, 32, 3743–3755. [Google Scholar] [CrossRef] [PubMed]
- Lalić, J.; PosavecMarjanović, M.; Palazzo, L.; Perina, D.; Sabljić, I.; Žaja, R.; Colby, T.; Pleše, B.; Halasz, M.; Jankevicius, G.; et al. Disruption of Macrodomain protein SCO6735 increases antibiotic production in Streptomyces coelicolor. J. Biol. Chem. 2016, 291, 23175–23187. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bonkowski, M.S.; Moniot, S.; Zhang, D.; Hubbard, B.P.; Ling, A.J.; Rajman, L.A.; Qin, B.; Lou, Z.; Gorbunova, V.; et al. A conserved NAD+ binding pocket that regulates protein-protein interactions during aging. Science 2017, 355, 1312–1317. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.M.; Thirawatananond, P.; Ong, S.E.; Gabelli, S.B.; Leung, A.K. Nudix hydrolases degrade protein-conjugated ADP-ribose. Sci. Rep. 2015, 5, 18271. [Google Scholar] [CrossRef] [PubMed]
- Bonfiglio, J.J.; Colby, T.; Matic, I. Mass spectrometry for serine ADP-ribosylation? Think o-glycosylation! Nucleic Acids Res. 2017, 45, 6259–6264. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, L.; Daniels, C.M.; Nettleship, J.E.; Rahman, N.; McPherson, R.L.; Ong, S.E.; Kato, K.; Nureki, O.; Leung, A.K.; Ahel, I. ENPP1 processes protein ADP-ribosylation in vitro. FEBS J. 2016, 283, 3371–3388. [Google Scholar] [CrossRef] [PubMed]
- Alderson, T. Ribonucleotide metabolism: Fresh approaches to viral and cancer chemotherapy. Biol. Rev. 1989, 64, 159–196. [Google Scholar] [CrossRef] [PubMed]
- Carmi-Levy, I.; Yannay-Cohen, N.; Kay, G.; Razin, E.; Nechushtan, H. Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells. Mol. Cell. Biol. 2008, 28, 5777–5784. [Google Scholar] [CrossRef] [PubMed]
- Monds, R.D.; Newell, P.D.; Wagner, J.C.; Schwartzman, J.A.; Lu, W.; Rabinowitz, J.D.; O’Toole, G.A. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J. Bacteriol. 2010, 192, 3011–3023. [Google Scholar] [CrossRef] [PubMed]
- Guranowski, A. Metabolism of diadenosine tetraphosphate (Ap4A) and related nucleotides in plants; review with historical and general perspective. Front. Biosci. 2004, 9, 1398–1411. [Google Scholar] [CrossRef] [PubMed]
- Despotovic, D.; Brandis, A.; Savidor, A.; Levin, Y.; Fumagalli, L.; Tawfik, D.S. Diadenosine tetraphosphate (Ap4A)—An E. coli alarmone or a damage metabolite? FEBS J. 2017, 284, 2194–2215. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Nechushtan, H.; Figov, N.; Razin, E. The function of lysyl-tRNA synthetase and Ap4A as signaling regulators of MITF activity in FcepsilonRI-activated mast cells. Immunity 2004, 20, 145–151. [Google Scholar] [CrossRef]
- Huebner, K.; Saldivar, J.C.; Sun, J.; Shibata, H.; Druck, T. Hits, Fhits and Nits: Beyond enzymatic function. Adv. Enzyme Regul. 2011, 51, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Brenner, C. Hint, Fhit, and GalT: Function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry 2002, 41, 9003–9014. [Google Scholar] [CrossRef] [PubMed]
- Motzik, A.; Amir, E.; Erlich, T.; Wang, J.; Kim, B.G.; Han, J.M.; Kim, J.H.; Nechushtan, H.; Guo, M.; Razin, E.; et al. Post-translational modification of HINT1 mediates activation of MITF transcriptional activity in human melanoma cells. Oncogene 2017, 36, 4732–4738. [Google Scholar] [CrossRef] [PubMed]
- Marriott, A.S.; Vasieva, O.; Fang, Y.; Copeland, N.A.; McLennan, A.G.; Jones, N.J. NUDT2 disruption elevates diadenosine tetraphosphate (Ap4A) and down-regulates immune response and cancer promotion genes. PLoS ONE 2016, 11, e0154674. [Google Scholar] [CrossRef] [PubMed]
- Crooke, A.; Guzman-Aranguez, A.; Carracedo, G.; de Lara, M.J.P.; Pintor, J. Understanding the presence and roles of Ap4A (Diadenosine tetraphosphate) in the eye. J. Ocul. Pharmacol. Ther. 2017, 33, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Reigada, D.; Navarro-Ruiz, R.M.; Caballero-López, M.J.; Del Águila, Á.; Muñoz-Galdeano, T.; Maza, R.M.; Nieto-Díaz, M. Diadenosine tetraphosphate (Ap4A) inhibits ATP-induced excitotoxicity: A neuroprotective strategy for traumatic spinal cord injury treatment. Purinergic Signal. 2017, 13, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, J.B.; Kjær, K.H.; Justesen, J.; Martensen, P.M. Enzyme assays for synthesis and degradation of 2-5As and other 2’-5’ oligonucleotides. BMC Biochem. 2015, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Tóth, B.; Iordanov, I.; Csanády, L. Ruling out pyridine dinucleotides as true TRPM2 channel activators reveals novel direct agonist ADP-ribose-2’-phosphate. J. Gen. Physiol. 2015, 145, 419–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühn, F.; Kühn, C.; Lückhoff, A. Different principles of ADP-ribose-mediated activation and opposite roles of the NUDT9 Homology Domain in the TRPM2 orthologs of man and sea anemone. Front. Physiol. 2017, 8, 879. [Google Scholar] [CrossRef] [PubMed]
- Di, A.; Kiya, T.; Gong, H.; Gao, X.; Malik, A.B. Role of the phagosomal redox-sensitive TRP channel TRPM2 in regulating bactericidal activity of macrophages. J. Cell Sci. 2017, 130, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Higashida, H.; Yokoyama, S.; Huang, J.J.; Liu, L.; Ma, W.J.; Akther, S.; Higashida, C.; Kikuchi, M.; Minabe, Y.; Munesue, T. Social memory, amnesia, and autism: Brain oxytocin secretion is regulated by NAD+ metabolites and single nucleotide polymorphisms of CD38. Neurochem. Int. 2012, 61, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Pereira, J.; Tarragó, M.G.; Chini, C.C.; Nin, V.; Escande, C.; Warner, G.M.; Puranik, A.S.; Schoon, R.A.; Reid, J.M.; Galina, A.; Chini, E.N. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016, 23, 1127–1139. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, A.R.; Larrick, J.W. The NAD+/PARP1/SIRT1 Axis in Aging. Rejuvenation Res. 2017, 20, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Dölle, C.; Rack, J.G.; Ziegler, M. NAD and ADP-ribose metabolism in mitochondria. FEBS J. 2013, 280, 3530–3541. [Google Scholar] [CrossRef] [PubMed]
- Diani-Moore, S.; Shoots, J.; Singh, R.; Zuk, J.B.; Rifkind, A.B. NAD+ loss, a new player in AhR biology: Prevention of thymus atrophy and hepatosteatosis by NAD+ repletion. Sci. Rep. 2017, 7, 2268. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, T.; Baur, J.A.; Perez, E.; Matsui, T.; Carmona, J.J.; Lamming, D.W.; Souza-Pinto, N.C.; Bohr, V.A.; Rosenzweig, A.; et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 2007, 130, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Martín-Guerrero, S.M.; Muñoz-Gámez, J.A.; Carrasco, M.-C.; Salmerón, J.; Martín-Estebané, M.; Cuadros, M.A.; Martín-Oliva, D. Poly(ADP-ribose)polymerases inhibitors prevent early mitochondrial fragmentation and hepatocyte cell death induced by H2O2. PLoS ONE 2017, 12, e0187130. [Google Scholar] [CrossRef] [PubMed]
- Liou, G.G.; Tanny, J.C.; Kruger, R.G.; Walz, T.; Moazed, D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 2005, 121, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Denu, J.M. Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 2010, 1804, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Billington, R.A.; Travelli, C.; Ercolano, E.; Galli, U.; Roman, C.B.; Grolla, A.A.; Canonico, P.L.; Condorelli, F.; Genazzani, A.A. Characterization of NAD uptake in mammalian cells. J. Biol. Chem. 2008, 283, 6367–6374. [Google Scholar] [CrossRef] [PubMed]
- Calcraft, P.J.; Ruas, M.; Pan, Z.; Cheng, X.; Arredouani, A.; Hao, X.; Tang, J.; Rietdorf, K.; Teboul, L.; Chuang, K.-T.; et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 2009, 459, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Virág, L.; Robaszkiewicz, A.; Rodriguez-Vargas, J.M.; Oliver, F.J. Poly(ADP-ribose) signaling in cell death. Mol. Asp. Med. 2013, 34, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Henderson, D.J.P.; Miranda, J.L.; Emerson, B.M. The ß-NAD salvage pathway and PKC-mediated signaling influence localized PARP-1 activity and CTCF poly(ADP) ribosylation. Oncotarget 2017, 8, 64698–64713. [Google Scholar] [CrossRef] [PubMed]
- Krietsch, J.; Rouleau, M.; Pic, É.; Ethier, C.; Dawson, T.M.; Dawson, V.L.; Masson, J.Y.; Poirier, G.G.; Gagné, J.P. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol. Asp. Med. 2013, 34, 1066–1087. [Google Scholar] [CrossRef] [PubMed]
- Till, S.; Ladurner, A.G. Sensing NAD metabolites through macrodomains. Front. Biosci. 2009, 14, 3246–3258. [Google Scholar] [CrossRef]
- Nakano, T.; Takahashi-Nakaguchi, A.; Yamamoto, M.; Watanabe, M. Pierisins and CARP-1: ADP-ribosylation of DNA by ARTCs in butterflies and shellfish. In Endogenous ADP-Ribosylation; Koch-Nolte, F., Ed.; Springer International Publishing: New York, NY, USA, 2015; pp. 127–149. [Google Scholar]
- Watanabe, M.; Kono, T.; Matsushima-Hibiya, Y.; Kanazawa, T.; Nishisaka, N.; Kishimoto, T.; Koyama, K.; Sugimura, T.; Wakabayashi, K. Molecular cloning of an apoptosis-inducing protein, pierisin, from cabbage butterfly: Possible involvement of ADP-ribosylation in its activity. Proc. Natl. Acad. Sci. USA 1999, 96, 10608–10613. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.S.; Citarelli, M.; Teotia, S. Functions of the poly(ADP-ribose) polymerase superfamily in plants. Cell. Mol. Life Sci. 2012, 69, 175–189. [Google Scholar] [CrossRef] [PubMed]
- Jaspers, P.; Overmyer, K.; Wrzaczek, M.; Vainonen, J.P.; Blomster, T.; Salojärvi, J.; Reddy, R.A.; Kangasjärvi, J. The RST and PARP-like domain containing SRO protein family: Analysis of protein structure, function and conservation in land plants. BMC Genom. 2010, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.S. Abiotic stress responses in plants: A focus on the SRO family. In Selected Plant Physiology Aspects; Montanaro, G., Ed.; InTech: Rijeka, Croatia, 2010; pp. 3–22. [Google Scholar]
- Adams-Phillips, L.; Briggs, A.G.; Bent, A.F. Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis thaliana to biotic stress. Plant Physiol. 2010, 152, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Rissel, D.; Heym, P.P.; Thor, K.; Brandt, W.; Wessjohann, L.A.; Peiter, E. No silver bullet—Canonical Poly(ADP-Ribose) Polymerases (PARPs) are no universal factors of biotic and biotic stress resistance of Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Liu, C.; Shan, L.; He, P. Protein ADP-ribosylation takes control in plant-bacterium interactions. PLoS Pathog. 2016, 12, e1005941. [Google Scholar] [CrossRef] [PubMed]
- Menke, F.L.H. Plants get on PAR with poly(ADP-ribosyl)ation. EMBO Rep. 2016, 17, 1677–1678. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Ma, S.; Chen, S.; Zhu, N.; Zhang, S.; Yu, B.; Yu, Y.; Le, B.; Chen, X.; Dinesh-Kumar, S.P.; et al. PARylation of the forkhead-associated domain protein DAWDLE regulates plant immunity. EMBO Rep. 2016, 17, 1799–1813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Mou, Z. Extracellular pyridine nucleotides induce PR gene expression and disease resistance in Arabidopsis. Plant J. 2009, 57, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhou, M.; Zhang, X.; Yao, J.; Zhang, Y.; Mou, Z. A lectin receptor kinase as a potential sensor for extracellular nicotinamide adenine dinucleotide in Arabidopsis thaliana. eLife 2017, 6, e25474. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; Bont, L.; Tcherkez, G.; Gakière, B. Not just a pawn on the board of plant-pathogen interaction. Plant Signal. Behav. 2013, 8, e22477. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Queval, G.; Gakiere, B. NAD(P) synthesis and pyridine nucleotide cycling in plants and their potential importance in stress conditions. J. Exp. Bot. 2006, 57, 1603–1620. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; Tcherkez, G.; Gakière, B. Pyridine nucleotides induce changes in cytosolic pools of calcium in Arabidopsis. Plant Signal. Behav. 2016, 11, e1249082. [Google Scholar] [CrossRef] [PubMed]
- Pétriacq, P.; de Bont, L.; Hager, J.; Didierlaurent, L.; Mauve, C.; Guérard, F.; Noctor, G.; Pelletier, S.; Renou, J.P.; Tcherkez, G.; Gakière, B. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J. 2012, 70, 650–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pétriacq, P.; Ton, J.; Patrit, O.; Tcherkez, G.; Gakière, B. NAD acts as an integral regulator of multiple defense layers. Plant Physiol. 2016, 172, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Miwa, A.; Sawada, Y.; Tamaoki, D.; Yokota Hirai, M.; Kimura, M.; Sato, K.; Nishiuchi, T. Nicotinamide mononucleotide and related metabolites induce disease resistance against fungal phytopathogens in Arabidopsis and barley. Sci. Rep. 2017, 7, 6389. [Google Scholar] [CrossRef] [PubMed]
- Adams-Phillips, L.; Wan, J.; Tan, X.; Dunning, F.M.; Meyers, B.C.; Michelmore, R.W.; Bent, A.F. Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol. Plant Microbe Interact. 2008, 21, 646–665. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Y.; Zhou, D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017, 45, 12241–12255. [Google Scholar] [CrossRef] [PubMed]
- Ashihara, H.; Ludwig, I.A.; Katahira, R.; Yokota, T.; Fujimura, T.; Crozier, A. Trigonelline and related nicotinic acid metabolites: Occurrence, biosynthesis, taxonomic considerations, and their roles in planta and in human health. Phytochem. Rev. 2015, 14, 765–798. [Google Scholar] [CrossRef]
- Ishikawa, K.; Ogawa, T.; Hirosue, E.; Nakayama, Y.; Harada, K.; Fukusaki, E.; Yoshimura, K.; Shigeoka, S. Modulation of the poly(ADP-ribosyl)ation reaction via the Arabidopsis ADP-ribose/NADH pyrophosphohydrolase, AtNUDX7, is involved in the response to oxidative stress. Plant Physiol. 2009, 151, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Yoshimura, K.; Harada, K.; Fukusaki, E.; Ogawa, T.; Tamoi, M.; Shigeoka, S. AtNUDX6, an ADP-ribose/NADH pyrophosphohydrolase in Arabidopsis, positively regulates NPR1-dependent salicylic acid signaling. Plant Physiol. 2010, 152, 2000–2012. [Google Scholar] [CrossRef] [PubMed]
- Shidore, T.; Broeckling, C.D.; Kirkwood, J.S.; Long, J.J.; Miao, J.; Zhao, B.; Leach, J.E.; Triplett, L.R. The effector AvrRxo1 phosphorylates NAD in planta. PLoS Pathog. 2017, 13, e1006442. [Google Scholar] [CrossRef] [PubMed]
- Luna, E.; van Hulten, M.; Zhang, Y.; Berkowitz, O.; López, A.; Pétriacq, P.; Sellwood, M.A.; Chen, B.; Burrell, M.; van de Meene, A.; et al. Plant perception of β-aminobutyric acid is mediated by an aspartyl-tRNAsynthetase. Nat. Chem. Biol. 2014, 10, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Glauser, G.; Mauch-Mani, B. The accumulation of β-aminobutyric acid is controlled by the plant’s immune system. Planta 2017. [Google Scholar] [CrossRef] [PubMed]
- Thevenet, D.; Pastor, V.; Baccelli, I.; Balmer, A.; Vallat, A.; Neier, R.; Glauser, G.; Mauch-Mani, B. The priming molecule β-aminobutyric acid is naturally present in plants and is induced by stress. New Phytol. 2017, 213, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Mauch-Mani, B. Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004, 38, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbacher, R.E.; Luna, E.; Ton, J. The discovery of the BABA receptor: Scientific implications and application potential. Front. Plant Sci. 2014, 5, 304. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poltronieri, P.; Čerekovic, N. Roles of Nicotinamide Adenine Dinucleotide (NAD+) in Biological Systems. Challenges 2018, 9, 3. https://doi.org/10.3390/challe9010003
Poltronieri P, Čerekovic N. Roles of Nicotinamide Adenine Dinucleotide (NAD+) in Biological Systems. Challenges. 2018; 9(1):3. https://doi.org/10.3390/challe9010003
Chicago/Turabian StylePoltronieri, Palmiro, and Nataša Čerekovic. 2018. "Roles of Nicotinamide Adenine Dinucleotide (NAD+) in Biological Systems" Challenges 9, no. 1: 3. https://doi.org/10.3390/challe9010003
APA StylePoltronieri, P., & Čerekovic, N. (2018). Roles of Nicotinamide Adenine Dinucleotide (NAD+) in Biological Systems. Challenges, 9(1), 3. https://doi.org/10.3390/challe9010003