The Development of a Defect Detection Model from the High-Resolution Images of a Sugarcane Plantation Using an Unmanned Aerial Vehicle
Abstract
:1. Introduction
2. Material and Method
2.1. Data Collection
2.2. Image Preprocessing
2.2.1. Shadow Detection
- Step 1: Convert the color image to LAB color space.
- Step 2: Calculate the mean of each color plane followed by the standard deviation of L plane.
- Step 3: Detect the shadow pixel by thresholding. The conditions for thresholding are shown in the Figure 5 and the algorithm is shown as a pseudo-code in Algorithm 1.
- Step 4: Divide the areas into 2 regions: the shadow regions and the non-shadow region regions . The shadow region has a pixel value of 0 (black) and the non-shadow region has a pixel value of 255 (white). The results from the shadow detection method are used as an input for shadow adjustment process.
| Algorithm 1 Pseudo-code description of a shadow detection. Input = UAV image. Output = Binary image |
| ( represents the shadow region and represents the non-shadow region) |
| 1: Procedure Shadow Detection (image) |
| 2: height ← Image height from image |
| 3: width ← Image width from image |
| 4: vertical scan at h: |
| 5: for x {0, height} do |
| 5: raster scan at w: |
| 6: for y {0, width} do |
| 7: If mean (A (x ,y)) + mean (B (x,y)) <= 256 |
| 8: If (L (x ,y) <= mean (L)) – standard deviation (L) /3 |
| 9: shadow region () |
| 10: Else |
| 11: non-shadow region () |
| 12: end if |
| 13: Else |
| 14: If pixel (L(x ,y)) < pixel(B (x ,y)) |
| 15: shadow region () |
| 16: Else |
| 17: non-shadow region () |
| 18: end if |
| 19: end if |
| 20: end for |
| 21: end for |
| 22: End procedure |
2.2.2. Shadow Adjustment
- Step 1: Calculate the light source of the shadow and non-shadow areas in the output image given by the shadow detection method. The light source value was calculated from Equations (1) and (2).where
- is the xy-summation of the shadow region.
- is the number of pixels on the shadow region.
- is the scale factor defined by R channel (KR), G channel (KG), B channel (KB).
- p is the weight of each gray value in the light source.
where- is the xy-summation of the non-shadow region.
- is the number of pixels on the non-shadow region.
- is the scale factor defined by R channel (KR), G channel (KG), B channel (KB).
- p is the weight of each gray value in the light source.
- Step 2: Determine the value of p that is suitable for adjusting the intensity of the shadow areas. Brightness, contrast, and average gradient of the shadow area were compared with the non-shadow areas. The appropriate p-values generated from the experiments is shown in Section 3.2.
- Step 3: Calculate the constant ratio of the light source values in both areas and proceed to adjust the intensity of the shadow area as in Equation (3).where
- is the new shadow region.
- / is the rate constant of shadow and non-shadow regions.
- is a shadow region.
2.3. Defect Detection Model Creation
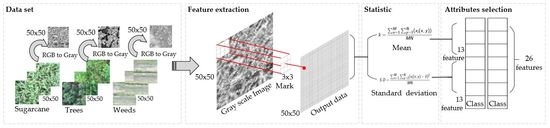
2.3.1. Feature Extraction
- Step 1: Convert RGB color image to grayscale format.
- Step 3: The statistical values such as mean and standard deviation are calculated from step 2. This process achieves 26 characteristics.
- Step 4: Select the features that are related to the class of the category using the WEKA program [29]. WEKA program uses the “Information Gain-Based Feature” selection method to find the significant features which is known as entropy. The features must pass the threshold value of more than 0.05. This value is selected using a T-score distribution method which separates the group of data obtained from entropy and eliminates those groups that have least relationship with the class (sugarcane, trees, and weeds). When the threshold value 0.05 was used as a condition, it gives the most significant features for the classification of sugarcane areas.
2.3.2. The Sugarcane Areas Classification Process
2.3.3. The Defect Areas Classification Process
2.3.4. Data Integration Process
2.4. Application Program Creation
3. Experiment Results
3.1. The Performance Testing for Shadow Detection
3.2. The Performance Testing for Shadow Adjustment
3.3. The Performance Testing for Sugarcane Areas Classification
3.3.1. The Experiment of Feature Selection
3.3.2. The Experiment of Sugarcane Area Classification Model Creation
3.4. The Performance Testing for Defect Areas Classification
4. Result and Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sahu, O. Assessment of Sugarcane Industry: Suitability for Production, Consumption, and Utilization. Ann. Agrar. Sci. 2018, 16, 389–395. [Google Scholar] [CrossRef]
- Office, Eastern Region Development Commission. Targeted Industries. Available online: https://www.eeco.or.th/en/content/targeted-industries (accessed on 1 December 2019).
- Terpitakpong, J. Sustainable Sugarcane Farm Management Guide; Sugarcane and Sugar Industry Information Group, Office of the Cane and Sugar Board Ministry of Industry: Bangkok, Thailand, 2017; pp. 8–15. [Google Scholar]
- Nopakhun, C.; Chocipanyo, D. Academic Leadership Strategies of Kamphaeng Phet Rajabhat University for 2018–2022 Year. Golden Teak Hum. Soc. Sci. J. (GTHJ) 2019, 25, 35–48. [Google Scholar]
- Panitphichetwong. Bundit, 24 June 2017.
- Geo-Informatics. The Information Office of Space Technology and Remote Sensing System. Geo-Informatics and Space Technology Office. Available online: https://gistda.or.th/main/th/node/976 (accessed on 19 June 2019).
- Yano, I. Weed Identification in Sugarcane Plantation through Images Taken from Remotely Piloted Aircraft (Rpa) and Knn Classifier. J. Food Nutr. Sci. 2017, 5, 211. [Google Scholar]
- Plants, Institute for Surveying and Monitoring for Planting of Narcotic. Knowledge Management of Unmanned Aircraft for Drug Trafficking Surveys. Office of the Narcotics Control Board. Available online: https://www.oncb.go.th/ncsmi/doc3/ (accessed on 19 June 2019).
- Luna, I. Mapping Crop Planting Quality in Sugarcane from Uav Imagery: A Pilot Study in Nicaragua. J. Remote Sens. 2016, 8, 50. [Google Scholar] [CrossRef] [Green Version]
- Girolamo-Neto, C.D.; Sanches, I.D.; Neves, A.K.; Prudente, V.H.; Körting, T.S.; Picoli, M.C. Assessment of Texture Features for Bermudagrass (Cynodon Dactylon) Detection in Sugarcane Plantations. Drones 2019, 3, 36. [Google Scholar] [CrossRef] [Green Version]
- Mink, R.; Dutta, A.; Peteinatos, G.G.; Sökefeld, M.; Engels, J.J.; Hahn, M.; Gerhards, R. Multi-Temporal Site-Specific Weed Control of Cirsium arvense (L.) Scop. And Rumex crispus L. in Maize and Sugar Beet Using Unmanned Aerial Vehicle Based Mapping. Agriculture 2018, 8, 65. [Google Scholar] [CrossRef] [Green Version]
- Matese, A.; Di Gennaro, S.F. Practical Applications of a Multisensor Uav Platform Based on Multispectral, Thermal and Rgb High Resolution Images in Precision Viticulture. Agriculture 2018, 8, 116. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Mishra, S.; Khanna, P. Precision Sugarcane Monitoring Using Svm Classifier. Procedia Comput. Sci. 2017, 122, 881–887. [Google Scholar] [CrossRef]
- Liu, B.; Shi, Y.; Duan, Y.; Wu, W. Uav-Based Crops Classification with Joint Features from Orthoimage and DSM Data. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2018, 42, 1023–1028. [Google Scholar]
- Sanches, G.M.; Duft, D.G.; Kölln, O.T.; Luciano, A.C.; De Castro, S.G.; Okuno, F.M.; Franco, H.C. The Potential for Rgb Images Obtained Using Unmanned Aerial Vehicle to Assess and Predict Yield in Sugarcane Fields. Int. J. Remote Sens. 2018, 39, 5402–5414. [Google Scholar] [CrossRef]
- Yano, I.H.; Alves, J.R.; Santiago, W.E.; Mederos, B.J. Identification of Weeds in Sugarcane Fields through Images Taken by Uav and Random Forest Classifier. IFAC-PapersOnLine 2016, 49, 415–420. [Google Scholar] [CrossRef]
- Oooka, K.; Oguchi, T. Estimation of Synchronization Patterns of Chaotic Systems in Cartesian Product Networks with Delay Couplings. Int. J. Bifurc. Chaos 2016, 26, 1630028. [Google Scholar] [CrossRef]
- Natarajan, R.; Subramanian, J.; Papageorgiou, E.I. Hybrid Learning of Fuzzy Cognitive Maps for Sugarcane Yield Classification. Comput. Electron. Agric. 2016, 127, 147–157. [Google Scholar] [CrossRef]
- Anoopa, S.; Dhanya, V.; Kizhakkethottam, J.J. Shadow Detection and Removal Using Tri-Class Based Thresholding and Shadow Matting Technique. Procedia Technol. 2016, 24, 1358–1365. [Google Scholar] [CrossRef] [Green Version]
- Hiary, H.; Zaghloul, R.; Al-Zoubi, M.D. Single-Image Shadow Detection Using Quaternion Cues. Comput. J. 2018, 61, 459–468. [Google Scholar] [CrossRef]
- Park, K.H.; Kim, J.H.; Kim, Y.H. Shadow Detection Using Chromaticity and Entropy in Colour Image. Int. J. Inf. Technol. Manag. 2018, 17, 44. [Google Scholar] [CrossRef]
- Chavolla, E.; Zaldivar, D.; Cuevas, E.; Perez, M.A. Color Spaces Advantages and Disadvantages in Image Color Clustering Segmentation; Springer: Cham, Switzerland, 2018; pp. 3–22. [Google Scholar]
- Suny, A.H.; Mithila, N.H. A Shadow Detection and Removal from a Single Image Using Lab Color Space. Int. J. Comput. Sci. Issues 2013, 10, 270. [Google Scholar]
- Xiao, Y.; Tsougenis, E.; Tang, C.K. Shadow Removal from Single Rgb-D Images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2014, Columbus, OH, USA, 23–28 June 2014. [Google Scholar]
- Shahtahmassebi, A.; Yang, N.; Wang, K.; Moore, N.; Shen, Z. Review of Shadow Detection and De-Shadowing Methods in Remote Sensing. Chin. Geogr. Sci. 2013, 23, 403–420. [Google Scholar] [CrossRef] [Green Version]
- Movia, A.; Beinat, A.; Crosilla, F. Shadow Detection and Removal in Rgb Vhr Images for Land Use Unsupervised Classification. ISPRS J. Photogram. Remote Sens. 2016, 119, 485–495. [Google Scholar] [CrossRef]
- Ye, Q.; Xie, H.; Xu, Q. Removing Shadows from High-Resolution Urban Aerial Images Based on Color Constancy. Int. Arch. Photogramm. Remote Sens. Spatial Inf. Sci. 2012, 39, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, R.C.; Faisal, Z. Digital Image Processing, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar]
- Janošcová, R. Mining Big Data in Weka; School of Management in Trenčín: Trenčín, Slovakia, 2016. [Google Scholar]
- Imandoust, S.B.; Bolandraftar, M. Application of K-Nearest Neighbor (Knn) Approach for Predicting Economic Events Theoretical Background. Int. J. Eng. Res. Appl. 2013, 3, 605–610. [Google Scholar]
- WorkWithColor.com. The Color Wheel. Available online: http://www.workwithcolor.com/the-color-wheel-0666.htm (accessed on 10 December 2019).
- Zunic, J.; Rosin, P.L. Measuring Shapes with Desired Convex Polygons. IEEE Trans. Pattern Anal. Mach. Intell. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, J.; Zhang, C.J.; Jiang, B.; Chen, J.; Song, J.; Liu, Z.; He, Z.; Wong, S.Y.; Fang, P.H.; Ming, W.K. Artificial Intelligence versus Clinicians in Disease Diagnosis: Systematic Review. JMIR Med. Inform. 2019, 7, e10010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cipolla, R.; Battiato, S.; Farinella, G.M. (Eds.) Registration and Recognition in Images and Videos; Springer: Berlin, Germany, 2014; Volume 532, pp. 38–39. [Google Scholar]

















| Factor | Details of the Factors |
|---|---|
| Varieties | LK-91-11, Khon Kaen 3, U Thong 11 |
| Ratoon | Ratoon 1, Ratoon 2, Ratoon 3 |
| Soil series | 3, 4, 5, 6, 7, 15, 17, 18, 21, 22, 23, 33, 35, 36, 38, 40, 44, 46, 48, 49, 55, 56 |
| Yield levels | Low, Medium, High |
| Category | Dataset 1 | Dataset 2 | ||
|---|---|---|---|---|
| Training and Validation Set | Testing Set | Training and Validation Set | Testing Set | |
| Sugarcanes | 5956 | 1449 | 5956 | 1449 |
| Trees | 2820 | 705 | - | - |
| weed | 3144 | 786 | 3144 | 786 |
| Total | 11,890 | 2940 | 9100 | 2235 |
| Region and p/Condition | Brightness | Contrast | Average Gradients | ||||||
|---|---|---|---|---|---|---|---|---|---|
| r | g | b | r | g | b | r | g | b | |
| shadow region | 46.60 | 67.54 | 63.82 | 21.63 | 22.62 | 21.02 | 3.29 | 3.85 | 3.52 |
| p = 1 | 130.28 | 146.91 | 126.16 | 52.53 | 46.05 | 45.43 | 77.33 | 77.25 | 74.22 |
| p = 2 | 130.05 | 146.84 | 126.27 | 52.45 | 46.03 | 45.47 | 77.16 | 77.20 | 74.30 |
| p = 3 | 129.87 | 146.76 | 126.31 | 52.39 | 46.00 | 45.50 | 77.02 | 77.14 | 74.35 |
| p = 4 | 129.74 | 146.69 | 126.36 | 52.35 | 45.99 | 45.50 | 76.92 | 77.10 | 74.35 |
| p = 5 | 129.66 | 146.64 | 126.39 | 52.32 | 45.97 | 45.52 | 76.85 | 77.07 | 74.38 |
| p = 6 | 129.59 | 146.60 | 126.42 | 52.30 | 45.96 | 45.53 | 76.80 | 77.04 | 74.40 |
| p = 7 | 129.55 | 146.57 | 126.45 | 52.29 | 45.96 | 45.54 | 76.76 | 77.02 | 74.42 |
| p = 8 | 129.51 | 146.55 | 126.48 | 52.28 | 45.95 | 45.55 | 76.74 | 77.01 | 74.43 |
| p = 9 | 129.48 | 146.54 | 126.50 | 52.27 | 45.95 | 45.55 | 76.71 | 77.00 | 74.45 |
| p = 10 | 129.46 | 146.53 | 126.52 | 52.26 | 45.95 | 45.56 | 76.69 | 76.99 | 74.46 |
| non shadow region | 130.32 | 146.92 | 126.15 | 51.89 | 45.75 | 45.71 | 71.45 | 73.17 | 70.84 |
| Attribute | Dataset 1 | Dataset 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 10 × 10 | 20 × 20 | 25 × 25 | 40 × 40 | 50 × 50 | 10 × 10 | 20 × 20 | 25 × 25 | 40 × 40 | 50 × 50 | |
| Mean1 | 0.027 | 0.047 | 0.056 | 0.074 | 0.083 | 0.026 | 0.038 | 0.045 | 0.061 | 0.070 |
| Mean2 | 0.033 | 0.055 | 0.066 | 0.085 | 0.100 | 0.030 | 0.046 | 0.055 | 0.068 | 0.077 |
| Mean3 | 0.056 | 0.084 | 0.090 | 0.115 | 0.123 | 0.048 | 0.064 | 0.072 | 0.085 | 0.096 |
| Mean4 | 0.057 | 0.087 | 0.099 | 0.120 | 0.127 | 0.049 | 0.067 | 0.076 | 0.094 | 0.102 |
| Mean5 | 0.068 | 0.103 | 0.116 | 0.138 | 0.145 | 0.057 | 0.081 | 0.093 | 0.104 | 0.115 |
| Mean6 | 0.068 | 0.104 | 0.115 | 0.137 | 0.146 | 0.057 | 0.084 | 0.094 | 0.106 | 0.117 |
| Mean7 | 0.065 | 0.107 | 0.121 | 0.149 | 0.156 | 0.055 | 0.088 | 0.099 | 0.117 | 0.124 |
| Mean8 | 0.069 | 0.108 | 0.122 | 0.148 | 0.160 | 0.057 | 0.088 | 0.099 | 0.118 | 0.126 |
| Mean9 | 0.048 | 0.076 | 0.086 | 0.105 | 0.120 | 0.041 | 0.063 | 0.068 | 0.082 | 0.093 |
| Mean10 | 0.054 | 0.029 | 0.019 | 0.005 | 0.004 | 0.080 | 0.056 | 0.041 | 0.017 | 0.014 |
| Mean11 | 0.048 | 0.020 | 0.015 | 0.008 | 0.011 | 0.074 | 0.037 | 0.030 | 0.023 | 0.025 |
| Mean12 | 0.058 | 0.023 | 0.013 | 0.005 | 0.008 | 0.094 | 0.059 | 0.047 | 0.028 | 0.023 |
| Mean13 | 0.046 | 0.017 | 0.009 | 0.005 | 0.008 | 0.076 | 0.038 | 0.030 | 0.014 | 0.018 |
| Std1 | 0.090 | 0.224 | 0.295 | 0.465 | 0.519 | 0.012 | 0.108 | 0.176 | 0.371 | 0.458 |
| Std2 | 0.099 | 0.262 | 0.340 | 0.497 | 0.548 | 0.016 | 0.148 | 0.228 | 0.430 | 0.513 |
| Std3 | 0.096 | 0.187 | 0.235 | 0.363 | 0.419 | 0.011 | 0.062 | 0.105 | 0.239 | 0.309 |
| Std4 | 0.116 | 0.238 | 0.295 | 0.432 | 0.481 | 0.023 | 0.113 | 0.173 | 0.332 | 0.402 |
| Std5 | 0.175 | 0.329 | 0.398 | 0.534 | 0.570 | 0.065 | 0.203 | 0.284 | 0.457 | 0.522 |
| Std6 | 0.147 | 0.264 | 0.325 | 0.469 | 0.521 | 0.040 | 0.132 | 0.192 | 0.360 | 0.438 |
| Std7 | 0.319 | 0.503 | 0.551 | 0.607 | 0.611 | 0.249 | 0.486 | 0.560 | 0.669 | 0.690 |
| Std8 | 0.197 | 0.363 | 0.431 | 0.559 | 0.592 | 0.090 | 0.252 | 0.337 | 0.513 | 0.571 |
| Std9 | 0.100 | 0.242 | 0.310 | 0.479 | 0.533 | 0.018 | 0.113 | 0.182 | 0.381 | 0.466 |
| Std10 | 0.100 | 0.221 | 0.287 | 0.448 | 0.504 | 0.017 | 0.096 | 0.158 | 0.348 | 0.438 |
| Std11 | 0.095 | 0.227 | 0.296 | 0.468 | 0.533 | 0.013 | 0.108 | 0.176 | 0.391 | 0.491 |
| Std12 | 0.105 | 0.255 | 0.332 | 0.507 | 0.566 | 0.021 | 0.138 | 0.219 | 0.445 | 0.536 |
| Std13 | 0.105 | 0.231 | 0.300 | 0.472 | 0.533 | 0.025 | 0.111 | 0.182 | 0.394 | 0.492 |
| Sub-Image Size/Dataset | Dataset 1 | Dataset 2 | Average | ||
|---|---|---|---|---|---|
| 10-Fold Cross-Validation | Testing Set | 10-Fold Cross-Validation | Testing Set | Accuracy | |
| 10 × 10 | 81.4945 | 77.9043 | 87.4945 | 86.1586 | 83.2629 |
| 20 × 20 | 86.6946 | 82.7921 | 92.8462 | 91.5462 | 88.4697 |
| 25 × 25 | 88.5235 | 83.3612 | 94.3187 | 92.4223 | 89.6564 |
| 40 × 40 | 90.5117 | 85.4704 | 96.0659 | 94.6124 | 91.6651 |
| 50 × 50 | 91.3842 | 85.6522 | 96.7473 | 95.0109 | 92.1986 |
| Performance Factors | Dataset 1 | Dataset 2 |
|---|---|---|
| Precision | 67.44% | 77.58% |
| Recall | 70.19% | 80.79% |
| F1-measure | 66.67% | 77.53% |
| Overall accuracy | 69.38% | 85.66% |
| Crop | Stage | Detection | OA | Classifier | Feature Used |
|---|---|---|---|---|---|
| Sugarcane [10] | mature | weed | 92.54% | RF | Multispectral |
| VI | |||||
| Texture | |||||
| Sugar beet [11] | tillering | weed | 96% | CHM | Multispectral |
| VI | |||||
| Maize [11] | tillering | weed | 80% | CHM | Multispectral |
| VI | |||||
| Vineyard [12] | mature | Missing plant | 80% | CWSI | Multispectral |
| NDVI | |||||
| Sugarcane [13] | mature | Leaf Disease | 96% | SVM | Spectral |
| Sensor Data | |||||
| Sugarcane [15] | tillering | Sugarcane Stalk | - | - | Spectral |
| GRVI | |||||
| Sugarcane [16] | tillering | weed | 82% | RF | Spectral |
| Texture | |||||
| Statistical | |||||
| Mean | |||||
| Average Deviation | |||||
| Standard Deviation | |||||
| Variance | |||||
| Kurtosis | |||||
| Skewness | |||||
| Maximum | |||||
| Minimum | |||||
| Sugarcane | mature | weed | 96.75% | KNN | Spectral |
| This paper | Texture | ||||
| Mean | |||||
| Standard deviation |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanut, B.; Riyamongkol, P. The Development of a Defect Detection Model from the High-Resolution Images of a Sugarcane Plantation Using an Unmanned Aerial Vehicle. Information 2020, 11, 136. https://doi.org/10.3390/info11030136
Tanut B, Riyamongkol P. The Development of a Defect Detection Model from the High-Resolution Images of a Sugarcane Plantation Using an Unmanned Aerial Vehicle. Information. 2020; 11(3):136. https://doi.org/10.3390/info11030136
Chicago/Turabian StyleTanut, Bhoomin, and Panomkhawn Riyamongkol. 2020. "The Development of a Defect Detection Model from the High-Resolution Images of a Sugarcane Plantation Using an Unmanned Aerial Vehicle" Information 11, no. 3: 136. https://doi.org/10.3390/info11030136
APA StyleTanut, B., & Riyamongkol, P. (2020). The Development of a Defect Detection Model from the High-Resolution Images of a Sugarcane Plantation Using an Unmanned Aerial Vehicle. Information, 11(3), 136. https://doi.org/10.3390/info11030136