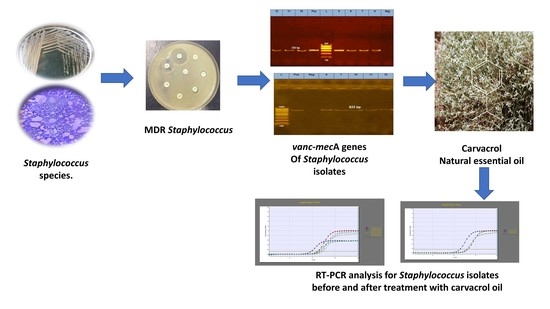
Carvacrol Essential Oil: A Natural Antibiotic against Zoonotic Multidrug-Resistant Staphylococcus Species Isolated from Diseased Livestock and Humans
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Staphylococcus Species
2.2. Identification and Prevalence of Different Staphylococcus spp. in Different Examined Samples
2.3. Haemolytic Activity and Biofilm Formation
2.4. Antimicrobial Susceptibility Testing
2.5. PCR Detection of Resistance- and Virulence-Associated Genes
2.6. Carvacrol EO Antibacterial Effect and MIC for Multidrug-Resistant Staphylococcus Isolates
2.7. Carvacrol EO Antimicrobial Susceptibility with MDR Staphylococcus Isolates
2.8. MDR Staphylococcus Isolates’ Real-Time PCR Analysis before and after Treatment with Carvacrol Oil
3. Discussion
4. Material and Methods
4.1. Samples
4.1.1. Animal Samples
4.1.2. Human Samples
4.2. Bacteriological Examination of the Collected Samples
4.3. Identification of Staphylococcus Isolates
4.4. Antimicrobial Susceptibility Profiles of the Staphylococcal Isolates
4.5. Phenotypic Detection of Slime Production (Biofilm Formation) on Congo Red Agar Medium
4.6. Polymerase Chain Reaction
4.7. Agar Dilution Method for Detection of the Antibacterial Effect Carvacrol Essential Oils on Staphylococcus Isolates
4.8. Phenotypic Effect of Carvacrol EO on Antimicrobial Susceptibility Profile of MDR Staphylococcus Isolates
4.9. SYBR Green RT-PCR on MDR Staphylococcus Isolates before and after Treatment with Carvacrol Oil
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Haag, A.F.; Fitzgerald, J.R.; Penadés, J.R. Staphylococcus aureus in Animals. Microbiol. Spectr. 2019, 7, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Carfora, V.; Caprioli, A.; Marri, N.; Sagrafoli, D.; Boselli, C.; Giacinti, G.; Giangolini, G.; Sorbara, L.; Dottarelli, S.; Battisti, A. Enterotoxin genes, enterotoxin production, and methicillin resistance in Staphylococcus aureus isolated from milk and dairy products in Central Italy. Int. Dairy J. 2015, 42, 12–15. [Google Scholar] [CrossRef] [Green Version]
- Wh, H.; Me, H.; Ha, E.; Sh, S. Characterization of antimicrobial resistant bacterial pathogens recovered from cases of bovine mastitis with special reference to Staphylococcus aureus. J. Vet. Med. Res. 2016, 23, 15–25. [Google Scholar]
- Seixas, R.V.D.; Bexiga, R.; Tavares, L.; Oliveira, M. Biofilm-formation by Staphylococcus aureus and Staphylococcus epidermidis isolates from subclinical mastitis in conditions mimicking the udder environment. Pol. J. Vet. Sci. 2015, 18, 787–792. [Google Scholar] [CrossRef] [Green Version]
- Todar, K.; Staphylococcus Aureus and Staphylococcal Disease. Todar’s Online Textbook of Bacteriology. 2008. Available online: http://www.textbookofbacteriology.net/staph.htm (accessed on 23 September 2021).
- Li, Z.; Stevens, D.L.; Hamilton, S.M.; Parimon, T.; Ma, Y.; Kearns, A.M.; Ellis, R.W.; Bryant, A.E. Fatal S. aureus hemorrhagic pneumonia: Genetic analysis of a unique clinical isolate producing both PVL and TSST-1. PLoS ONE 2011, 6, e27246. [Google Scholar] [CrossRef]
- Bochniarz, M.; Dzięgiel, B.; Nowaczek, A.; Wawron, W.; Dąbrowski, R.; Szczubiał, M.; Winiarczyk, S. Factors responsible for subclinical mastitis in cows caused by Staphylococcus chromogenes and its susceptibility to antibiotics based on bap, fnbA, eno, mecA, tetK, and ermA genes. J. Dairy Sci. 2016, 99, 9514–9520. [Google Scholar] [CrossRef] [Green Version]
- Jakeen, K.E.; Noha, E.A.; Alaa, G.; Mahmoud, D.; Hussein, M.G.; Sherif, A.O.; Ahmed, S. Emerging of coagulase negative Staphylococci as a cause of mastitis in dairy animals: An environmental hazard I. Int. J. Vet. Sci. Med. 2013, 1, 74–78. [Google Scholar] [CrossRef] [Green Version]
- Soares, L.C.; Pereira, I.A.; Pribul, B.R.; Oliva, M.S.; Coelho, S.M.O.; Souza, M. Antimicrobial resistance and detection of mecA and blaZ genes in coagulase-negative Staphylococcus isolated from bovine mastitis. Pesqui. Vet. Bras. 2012, 32, 692–696. [Google Scholar] [CrossRef] [Green Version]
- Klimiene, I.; Virgailis, M.; Pavilonis, A.; Siugzdiniene, R.; Mockeliunas, R.; Ruzauskas, M. Phenotypical and genotypical antimicrobial resistance of coagulase-negative Staphylococci isolated from cow mastitis. Pol. J. Vet. Sci. 2016, 19, 639–646. [Google Scholar] [CrossRef] [Green Version]
- Argudín, M.Á.; Mendoza, M.C.; Rodicio, M.R. Food poisoning and Staphylococcus aureus enterotoxins. Toxins 2010, 2, 1751–1773. [Google Scholar] [CrossRef]
- Bochniarz, M.; Wawron, W.; Szczubial, M. Production of slime by coagulase-negative Staphylococci (CNS) isolated from clinical and subclinical mastitis in cows. Pol. J. Vet. Sci. 2014, 17, 447–452. [Google Scholar] [CrossRef]
- Cree, R.G.A.; Aleljung, P.; Paulsson, M.; Witte, W.; Noble, W.C.; Ljungh, A.; Wadström, T. Cell surface hydrophobicity and adherence to extra–cellular matrix proteins in two collections of methicillin–resistant Staphylococcus aureus. Epidemiol. Infect. 1994, 112, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Agarwal, A.; Verma, R.K. Cefoxitin disc diffusion test for detection of meticillin-resistant Staphylococci. J. Med. Microbiol. 2008, 57, 957–961. [Google Scholar] [CrossRef]
- Nasr, R.A.; AbuShady, H.M.; Hussein, H.S. Biofilm formation and presence of icaAD gene in clinical isolates of Staphylococci. Egypt. J. Med. Hum. Genet. 2012, 13, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Abed, A.H.; Menshawy, A.; Zeinhom, M.; Hossain, D.; Khalifa, E.; Wareth, G.; Awad, M.F. Subclinical Mastitis in Selected Bovine Dairy Herds in North Upper Egypt: Assessment of Prevalence, Causative Bacterial Pathogens, Antimicrobial Resistance and Virulence-Associated Genes. Microorganisms 2021, 9, 1175. [Google Scholar] [CrossRef]
- Abed, A.H.; Al Sayed, R.A.; Atia, A.A. Genotyping of β-lactams resistant Staphylococci isolated from bovine subclinical mastitis. Beni-Suef Univ. J. Basic Appl. Sci. 2018, 7, 499–504. [Google Scholar] [CrossRef]
- Azhar, A.; Rasool, S.; Haque, A.; Shan, S.; Saeed, M.; Ehsan, B.; Haque, A. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J. Med. Microbiol. 2017, 66, 1328–1331. [Google Scholar] [CrossRef]
- McGuinness, W.A.; Malachowa, N.; DeLeo, F.R. Focus: Infectious diseases: Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 2017, 90, 269. [Google Scholar]
- Cong, Y.; Yang, S.; Rao, X. Vancomycin resistant Staphylococcus aureus infections: A review of case updating and clinical features. J. Adv. Res. 2020, 21, 169–176. [Google Scholar] [CrossRef]
- World Health Organization. Tackling Antibiotic Resistance from a Food Safety Perspective in Europe; World Health Organization, Regional Office for Europe: Moscow, Russia, 2011. [Google Scholar]
- Radwan, I.A.; Abed, A.H.; Abd Al-Wanis, S.A.; Abd El-Aziz, G.G.; El-Shemy, A. Antibacterial effect of cinnamon and oreganium oils on multidrug resistant Escherichia coli and Salmonellae isolated from broiler chickens. J. Egypt. Vet. Med Assoc. 2016, 76, 169–186. [Google Scholar]
- Lopez-Romero, J.C.; González-Ríos, H.; Borges, A.; Simões, M. Antibacterial effects and mode of action of selected essential oils components against Escherichia coli and Staphylococcus aureus. Evid.-Based Complement. Altern. Med. 2015, 2015, 795435. [Google Scholar] [CrossRef] [Green Version]
- Torky, H.A.; Abu Tabeikh, S.M. Incidence of Coagulase Negative Staphylococcus Isolated from Mastitis Cows and Human Contact. Alex. J. Vet. Sci. 2016, 51, 112–117. [Google Scholar]
- Radwan, I.A.H.; Shehata, A.A.E.; Abdel-Ghani, A.E.; Abdullah, M.M.; Abdraboh, A.A.M. Phenotypic and genotypic diversity of Staphylococcus aureus isolated from livestock and human. Glob. Vet. 2015, 14, 274–281. [Google Scholar]
- Pantosti, A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Front. Microbiol. 2012, 3, 127. [Google Scholar] [CrossRef] [Green Version]
- Roberts, M.C.; Garland-Lewis, G.; Trufan, S.; Meschke, S.J.; Fowler, H.; Shean, R.C.; Greninger, A.L.; Rabinowitz, P.M. Distribution of Staphylococcus species in dairy cows, workers and shared farm environments. FEMS Microbiol. Lett. 2018, 365, fny146. [Google Scholar] [CrossRef]
- El-Seedy, F.R.; Radwan, I.A.; Hassan, W.H.; Shehata, A. Coagulase Negative Staphylococci as an emerging cause of bovine mastitis: Prevalence, antimicrobial resistance and biofilm formation. J. Vet. Med. Res. 2017, 24, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Moraveji, Z.; Tabatabaei, M.; Aski, H.S.; Khoshbakht, R. Characterization of hemolysins of Staphylococcus strains isolated from human and bovine, southern Iran. Iran. J. Vet. Res. 2014, 15, 326. [Google Scholar]
- Tremblay, Y.D.N.; Lamarche, D.; Chever, P.; Haine, D.; Messier, S.; Jacques, M. Characterization of the ability of coagulase-negative Staphylococci isolated from the milk of Canadian farms to form biofilms. J. Dairy Sci. 2013, 96, 234–246. [Google Scholar] [CrossRef] [Green Version]
- Osman, K.M.; Abd El-Razik, K.A.; Marie, H.S.H.; Arafa, A. Relevance of biofilm formation and virulence of different species of coagulase-negative Staphylococci to public health. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 2009–2016. [Google Scholar] [CrossRef]
- Hou, W.; Sun, X.; Wang, Z.; Zhang, Y. Biofilm-forming capacity of Staphylococcus epidermidis, Staphylococcus aureus, and Pseudomonas aeruginosa from ocular infections. Investig. Ophthalmol. Vis. Sci. 2012, 53, 5624–5631. [Google Scholar] [CrossRef] [Green Version]
- Murugan, K.; Usha, M.; Malathi, P.; Al-Sohaibani, A.S.; Chandrasekaran, M. Biofilm forming multi drug resistant Staphylococcus spp. among patients with conjunctivitis. Pol. J. Microbiol. 2010, 59, 233. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017, 8, 511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajala-Schultz, P.J.; Torres, A.H.; Degraves, F.J.; Gebreyes, W.A.; Patchanee, P. Antimicrobial resistance and genotypic characterization of coagulase-negative Staphylococci over the dry period. Vet. Microbiol. 2009, 134, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Diversity and antimicrobial susceptibility profiling of Staphylococci isolated from bovine mastitis cases and close human contacts. J. Dairy Sci. 2015, 98, 6256–6269. [Google Scholar] [CrossRef]
- Holmes, M.A.; Zadoks, R.N. Methicillin resistant S. aureus in human bovine mastitis. J. Mammary Gland Biol. Neoplasia 2011, 16, 373–382. [Google Scholar] [CrossRef]
- Werner, G.; Strommenger, B.; Witte, W. Acquired Vancomycin Resistance in Clinically Relevant Pathogens; Robert Koch Institute, Wernigerode Branch: Wernigerode, Germany, 2008. [Google Scholar]
- Zmantar, T.; Kouidhi, B.; Miladi, H.; Bakhrouf, A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative Staphylococci. BMC Res. Notes 2011, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Faezi, N.A.; Hasani, A.; Soltani, E.; Valizadeh, V.; Hasani, A.; Khabbaz, A.; Rezaee, M.A.; Varschochi, M. Plausible challenges of methicillin and clindamycin resistance detection in Staphylococcus aureus. Gene Rep. 2021, 24, 101179. [Google Scholar] [CrossRef]
- Farzana, K.; Rashid, Z.; Akhtar, N.; Sattar, A.; Khan, J.A.; Nasir, B. Nasal carriage of Staphylococci in health care workers: Antimicrobial susceptibility profile. Pak. J. Pharm. Sci. 2008, 21, 290–294. [Google Scholar]
- Timsina, R.; Shrestha, U.; Singh, A.; Timalsina, B. Inducible clindamycin resistance and erm genes in Staphylococcus aureus in school children in Kathmandu, Nepal. Future Sci. OA 2020, 7, FSO361. [Google Scholar] [CrossRef]
- Salgueiro, V.; Manageiro, V.; Bandarra, N.M.; Ferreira, E.; Clemente, L.; Caniça, M. Genetic Relatedness and Diversity of Staphylococcus aureus from Different Reservoirs: Humans and Animals of Livestock, Poultry, Zoo, and Aquaculture. Microorganisms 2020, 8, 1345. [Google Scholar] [CrossRef]
- Baum, C.; Haslinger-Loffler, B.; Westh, H.; Boye, K.; Peters, G.; Neumann, C.; Kahl, B.C. Non-spa-typeable clinical Staphylococcus aureus strains are naturally occurring protein A mutants. J. Clin. Microbiol. 2009, 47, 3624–3629. [Google Scholar] [CrossRef] [Green Version]
- Votintseva, A.A.; Fung, R.; Miller, R.R.; Knox, K.; Godwin, H.; Wyllie, D.H.; Bowden, R.; Crook, D.W.; Walker, A.S. Prevalence of protein A (spa) mutants in the community and hospitals in Oxfordshire. BMC Microbiol. 2014, 14, 63. [Google Scholar] [CrossRef] [Green Version]
- Peacock, S.J.; Moore, C.E.; Justice, A.; Kantzanou, M.; Story, L.; Mackie, K.; O’Neill, G.; Day, N.P. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 2002, 70, 4987–4996. [Google Scholar] [CrossRef] [Green Version]
- Koreen, L.; Ramaswamy, S.V.; Graviss, E.A.; Naidich, S.; Musser, J.M.; Kreiswirth, B.N. Spa typing method for discriminating among Staphylococcus aureus isolates: Implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 2004, 42, 792–799. [Google Scholar] [CrossRef] [Green Version]
- Brignoli, T.; Manetti, A.G.O.; Rosini, R.; Haag, A.F.; Scarlato, V.; Bagnoli, F.; Delany, I. Absence of Protein A Expression Is Associated With Higher Capsule Production in Staphylococcal Isolates. Front. Microbiol. 2019, 10, 863. [Google Scholar] [CrossRef]
- Bnyan, I.A.; Abid, A.T.; Obied, H.N. Antibacterial activity of carvacrol against different types of bacteria. J. Nat. Sci. Res. 2014, 4, 13–17. [Google Scholar]
- Memar, M.Y.; Raei, P.; Alizadeh, N.; Aghdam, M.A.; Kafil, H.S. Carvacrol and thymol: Strong antimicrobial agents against resistant isolates. Rev. Med. Microbiol. 2017, 28, 63–68. [Google Scholar] [CrossRef]
- Khoshbakht, T.; Karami, A.; Tahmasebi, A.; Maggi, F. The Variability of Thymol and Carvacrol Contents Reveals the Level of Antibacterial Activity of the Essential Oils from Different Accessions of Oliveria decumbens. Antibiotics 2020, 9, 409. [Google Scholar] [CrossRef]
- Zakaria Nabti, L.; Sahli, F.; Laouar, H.; Olowo-Okere, A.; Nkuimi Wandjou, J.G.; Maggi, F. Chemical composition and antibacterial activity of essential oils from the Algerian endemic Origanum glandulosum Desf. against multidrug-resistant uropathogenic E. coli isolates. Antibiotics 2020, 9, 29. [Google Scholar] [CrossRef] [Green Version]
- De Souza, G.H.d.A.; dos Santos Radai, J.A.; Mattos Vaz, M.S.; Esther da Silva, K.; Fraga, T.L.; Barbosa, L.S.; Simionatto, S. In vitro and in vivo antibacterial activity assays of carvacrol: A candidate for development of innovative treatments against KPC-producing Klebsiella pneumoniae. PLoS ONE 2021, 16, e0246003. [Google Scholar] [CrossRef]
- Wijesundara, N.M.; Lee, S.F.; Cheng, Z.; Davidson, R.; Rupasinghe, H.P.V. Carvacrol exhibits rapid bactericidal activity against Streptococcus pyogenes through cell membrane damage. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Schalm, O.W.; Carroll, J.E.; Jaire, N.C. Bovine Mastitis, 1st ed.; Lea and Febiger: Philadelphia, PA, USA, 1971; pp. 80–89. [Google Scholar]
- Quinn, P.J.; Markey, B.K.; Leonard, F.C.; Hartigan, P.; Fanning, S.; Fitzpatrick, E. Veterinary Microbiology and Microbial Disease, 2nd ed.; JohnWiley & Sons: Hoboken, NJ, USA, 2011; pp. 1–928. [Google Scholar]
- Waller, K.P.; Aspán, A.; Nyman, A.; Persson, Y.; Andersson, U.G. CNS species and antimicrobial resistance in clinical and subclinical bovine mastitis. Vet. Microbiol. 2011, 152, 112–116. [Google Scholar] [CrossRef] [Green Version]
- NMC (National Mastitis Council). Laboratory Handbook on Bovine Mastitis, 3rd ed.; National Mastitis Council Inc.: Madison, WI, USA, 2017; p. 147. [Google Scholar]
- CLSI (Clinical and Laboratory Standards Institute). Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Clinical and Laboratory Standards Institute. M100: Wayne, PA, USA, 2018. [Google Scholar]
- Chandran, A.; Hatha, A.A.M.; Varghese, S.; Sheeja, K.M. Prevalence of multiple drug-resistant Escherichia coli serotypes in a tropical estuary, India. Microbes Environ. 2008, 23, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Mason, W.J.; Blevins, J.S.; Beenken, K.; Wibowo, N.; Ojha, N.; Smeltzer, M.S. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 2001, 39, 3332–3338. [Google Scholar] [CrossRef] [Green Version]
- McClure, J.A.; Conly, J.M.; Lau, V.; Elsayed, S.; Louie, T.; Hutchins, W.; Zhang, K. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from-resistant Staphylococci. J. Clin. Microbiol. 2006, 44, 1141–1144. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.; Uhl, J.R.; Kohner, P.; Hopkins, M.K.; Cockerill, F.R. Multiplex PCR detection of vanA, vanB, vanC-1, and vanC-2/3 genes in enterococci. J. Clin. Microbiol. 1997, 35, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Kariyama, R.; Mitsuhata, R.; Chow, J.W.; Clewell, D.B.; Kumon, H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 2000, 38, 3092–3095. [Google Scholar] [CrossRef] [Green Version]
- Schlegelova, J.; Vlkova, H.; Babak, V.; Holasova, M.; Jaglic, Z.; Stosova, T.; Sauer, P. Resistance to erythromycin of Staphylococcus spp. isolates from the food chain. Vet. Med. 2008, 53, 307–314. [Google Scholar] [CrossRef]
- Fei, W.; Hongjun, Y.; Hong-bin, H.; Changfa, W.; Yundong, G.; Qifeng, Z.; Wiaohong, W.; Yanjun, Z. Study on the hemolysin phenotype and the genetype distribution of Staphylococcus aureus caused bovine mastitis in Shandong dairy farms. Int. J. Appl. Res. Vet. Med. 2011, 9, 416–421. [Google Scholar]
- Ciftci, A.; Findik, A.; Onuk, E.E.; Savasan, S. Detection of methicillin resistance and slime factor production of Staphylococcus aureus in bovine mastitis. Braz. J. Microbiol. 2009, 40, 254–261. [Google Scholar] [CrossRef] [Green Version]
- Wada, M.; Lkhagvadorj, E.; Bian, L.; Wang, C.; Chiba, Y.; Nagata, S.; Shimizu, T.; Yamashiro, Y.; Asahara, T.; Nomoto, K. Quantitative reverse transcription-PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 2010, 108, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, T.A.; Khalid, A.E.; Saadabi, A.M. PCR detection of staphylococcal enterotoxin A and B genes in Staphylococcus aureus isolated from salted fermented fish. Microbiol. J. 2014, 4, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Mehrtra, M.; Wang, G.; Johnson, W.M. Multiplex PCR for Detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J. Clin. Microbiol. 2000, 38, 1032–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radwan, I.A.; Abed, A.H.; Abdallah, A.S. Antifungal effect of carvacrol on fungal pathogens isolated from broiler chickens. Assiut Vet. Med. J. 2018, 64, 11–17. [Google Scholar] [CrossRef]
- Yuan, J.S.; Reed, A.; Chen, F.; Stewart, C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinform. 2006, 7, 85. [Google Scholar] [CrossRef] [Green Version]






| Samples | No. of Samples | Positive Bacterial Isolation | ||
|---|---|---|---|---|
| No. | % | |||
| Cow milk | 100 | 75 | 75 | |
| Sheep abscess | 25 | 13 | 52 | |
| Human samples | Wounds and abscesses | 30 | 22 | 73.3 |
| Dermal lesions | 33 | 24 | 72.7 | |
| Tracheal cavity | 28 | 20 | 71.4 | |
| Total human samples | 91 | 66 | 72.5 | |
| Overall total | 216 | 154 | 71.3 | |
| Bacterial Isolates | Milk Isolates | Sheep Isolates | Human Isolates | Overall Total No. of Isolates | ||||
|---|---|---|---|---|---|---|---|---|
| No | % | No | % | No | % | No | %* | |
| S. aureus | 35 | 46.7 | 7 | 53.8 | 24 | 36.4 | 66 | 42.9 |
| S schleiferi | 27 | 36 | 2 | 15.4 | 10 | 15.2 | 39 | 25.3 |
| S. intermedius | 8 | 10.7 | 2 | 15.4 | 9 | 13.6 | 19 | 12.3 |
| S. xylosus | 2 | 2.7 | 1 | 7.7 | 15 | 22.7 | 18 | 11.7 |
| S. haemolyticus | 2 | 2.7 | 0 | 0 | 5 | 7.6 | 7 | 4.5 |
| S. epidermidis | 1 | 1.3 | 1 | 7.7 | 1 | 1.5 | 3 | 1.9 |
| S. aurecularis | 0 | 0 | 0 | 0 | 2 | 3.0 | 2 | 1.3 |
| Total No. of isolates | 75 | 100 | 13 | 100 | 66 | 100 | 154 | 100 |
| Bacterial Isolates | No. of Tested Isolates | Carvacrol Oil Concentration | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1% | 0.05% | 0.04% | 0.03% | 0.02% | 0.01% | ||||||||
| No | % | No | % | No | % | No | % | No | % | No | % | ||
| S. aureus | 34 | 34 | 100 | 34 | 100 | 34 | 100 | 25 | 100 | 0 | 0 | 0 | 0 |
| S. schleiferi | 16 | 16 | 100 | 16 | 100 | 16 | 100 | 12 | 100 | 0 | 0 | 0 | 0 |
| S. intermedius | 8 | 8 | 100 | 8 | 100 | 8 | 100 | 6 | 100 | 0 | 0 | 0 | 0 |
| S. xylosus | 9 | 9 | 100 | 9 | 100 | 9 | 100 | 6 | 100 | 0 | 0 | 0 | 0 |
| S. haemolyticus | 4 | 4 | 100 | 4 | 100 | 4 | 100 | 4 | 100 | 0 | 0 | 0 | 0 |
| S. epidermidis | 1 | 1 | 100 | 1 | 100 | 1 | 100 | 1 | 100 | 0 | 0 | 0 | 0 |
| Total isolates | 72 | 72 | 100 | 72 | 100 | 72 | 100 | 54 | 75 | 0 | 0 | 0 | 0 |
| Total non-affected | 0 | 0 | 0 | 0 | 0 | 0 | 18 | 25 | 72 | 100 | 72 | 100 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abed, A.H.; Hegazy, E.F.; Omar, S.A.; Abd El-Baky, R.M.; El-Beih, A.A.; Al-Emam, A.; Menshawy, A.M.S.; Khalifa, E. Carvacrol Essential Oil: A Natural Antibiotic against Zoonotic Multidrug-Resistant Staphylococcus Species Isolated from Diseased Livestock and Humans. Antibiotics 2021, 10, 1328. https://doi.org/10.3390/antibiotics10111328
Abed AH, Hegazy EF, Omar SA, Abd El-Baky RM, El-Beih AA, Al-Emam A, Menshawy AMS, Khalifa E. Carvacrol Essential Oil: A Natural Antibiotic against Zoonotic Multidrug-Resistant Staphylococcus Species Isolated from Diseased Livestock and Humans. Antibiotics. 2021; 10(11):1328. https://doi.org/10.3390/antibiotics10111328
Chicago/Turabian StyleAbed, Ahmed H., Esraa F. Hegazy, Sherif A. Omar, Rehab M. Abd El-Baky, Ahmed A. El-Beih, Ahmed Al-Emam, Ahmed M. S. Menshawy, and Eman Khalifa. 2021. "Carvacrol Essential Oil: A Natural Antibiotic against Zoonotic Multidrug-Resistant Staphylococcus Species Isolated from Diseased Livestock and Humans" Antibiotics 10, no. 11: 1328. https://doi.org/10.3390/antibiotics10111328
APA StyleAbed, A. H., Hegazy, E. F., Omar, S. A., Abd El-Baky, R. M., El-Beih, A. A., Al-Emam, A., Menshawy, A. M. S., & Khalifa, E. (2021). Carvacrol Essential Oil: A Natural Antibiotic against Zoonotic Multidrug-Resistant Staphylococcus Species Isolated from Diseased Livestock and Humans. Antibiotics, 10(11), 1328. https://doi.org/10.3390/antibiotics10111328