Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa
Abstract
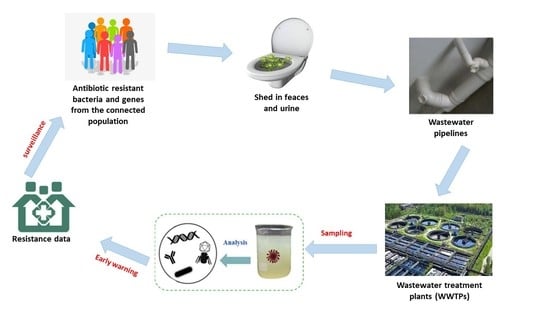
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Sample Collection and Processing
2.3. Selection of Antibiotic Resistance Genes
2.4. Optimization of Polymerase Chain Reaction Conditions
2.5. Optimization of ddPCR for Detection and Quantification of Tuberculosis-Resistance Genes in Wastewater
2.6. Statistical Analysis
3. Results
3.1. Detection of Genes Associated with Resistance to Drugs Used in TB Treatment Regimen Using Conventional PCR
3.2. Abundance of Antimicrobial Resistance Genes in Untreated and Treated Wastewater
3.2.1. Concentration of ARGs in the WWTPs
3.2.2. Variation in the Concentration of ARGs Responsible for Resistance to First- and Second-Line TB-Treatment Drugs and Other Related Drugs
3.3. Reduction in Antimicrobial Resistance Genes during Wastewater Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. WHO Treatment Guidelines for Drug-Resistant Tuberculosis; World Health Organization: Geneva, Switzerland, 2016. [Google Scholar]
- Kairigo, P.; Ngumba, E.; Sundberg, L.R.; Gachanja, A.; Tuhkanen, T. Occurrence of antibiotics and risk of antibiotic resistance evolution in selected Kenyan wastewaters, surface waters and sediments. Sci. Total Environ. 2020, 720, 137–580. [Google Scholar] [CrossRef]
- Varela, A.R.; André, S.; Nunes, O.C.; Manaia, C.M. Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system. Water Res. 2014, 54, 327–336. [Google Scholar] [CrossRef]
- Suthar, A.B.; Moonan, P.K.; Alexander, H.L. Towards national systems for continuous surveillance of antimicrobial resistance: Lessons from tuberculosis. PLoS Med. 2018, 15, e1002658. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, H.A. Profiling the host immune response to tuberculosis vaccines. Vaccine 2015, 33, 5313–5315. [Google Scholar] [CrossRef]
- Wang, H.; Edwards, M.; Falkinham III, J.O.; Pruden, A. Molecular survey of the occurrence of Legionella spp., Mycobacterium spp., Pseudomonas aeruginosa, and amoeba hosts in two chloraminated drinking water distribution systems. Appl. Environ. Microbiol. 2012, 78, 6285–6294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, H.; Dickson-Hall, L.; Ndjeka, N.; van’t Hoog, A.; Grant, A.; Cobelens, F.; Stevens, W.; Nicol, M. Delays and loss to follow-up before treatment of drug-resistant tuberculosis following implementation of Xpert MTB/RIF in South Africa: A retrospective cohort study. PLoS Med. 2017, 14, e1002238. [Google Scholar] [CrossRef]
- World Health Organization. WHO Global Tuberculosis Report 2018; Available online: https://www.who.int/tb/publications/global_report/gtbr2018_main_text_28Feb2019.pdf (accessed on 27 October 2021).
- Nnadozie, C.F.; Kumari, S.; Bux, F. Status of pathogens, antibiotic resistance genes and antibiotic residues in wastewater treatment systems. Rev. Environ. Sci. Bio/Technol. 2017, 16, 491–515. [Google Scholar] [CrossRef]
- Nguyen, Q.H. Genetic Determinants, and Evolution of Drug Resistance in Mycobacterium Tuberculosis in Vietnam: Toward New Diagnostic Tools. Ph.D. Thesis, Université Montpellier, Montpellier, France, 2016. [Google Scholar]
- Koch, A.; Cox, H.; Mizrahi, V. Drug-resistant tuberculosis: Challenges and opportunities for diagnosis and treatment. Curr. Opin. Pharmacol. 2018, 42, 7–15. [Google Scholar] [CrossRef]
- Ndjeka, N.; Hughes, J.; Reuter, A.; Conradie, F.; Enwerem, M.; Ferreira, H.; Ismail, N.; Kock, Y.; Master, I.; Meintjes, G.; et al. Implementing novel regimens for drug-resistant TB in South Africa: What can the world learn? Int. J. Tuberc. Lung Dis. 2020, 24, 1073–1080. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, L.H. Antimicrobial resistance: A One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 2017, 111, 255–260. [Google Scholar] [CrossRef]
- Sirgel, F.A.; Tait, M.; Warren, R.M.; Streicher, E.M.; Böttger, E.C.; Van Helden, P.D.; Gey van Pittius, N.C.; Coetzee, G.; Hoosain, E.Y.; Chabula-Nxiweni, M.; et al. Mutations in the rrs A1401G gene and phenotypic resistance to amikacin and capreomycin in Mycobacterium tuberculosis. Microb. Drug Resist. 2012, 18, 193–197. [Google Scholar] [CrossRef] [Green Version]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabri, N.A.; Schmitt, H.; Van der Zaan, B.; Gerritsen, H.W.; Zuidema, T.; Rijnaarts, H.H.; Langenhoff, A.A. Prevalence of antibiotics and antibiotic resistance genes in a wastewater effluent-receiving river in the Netherlands. J. Environ. Chem. Eng. 2020, 8, 102245. [Google Scholar] [CrossRef]
- Butkovskyi, A.; Leal, L.H.; Rijnaarts, H.H.M.; Zeeman, G. Fate of pharmaceuticals in full-scale source separated sanitation system. Water Res. 2015, 85, 384–392. [Google Scholar] [CrossRef]
- Nanninga, T.A.; Bisschops, I.; López, E.; Martínez-Ruiz, J.L.; Murillo, D.; Essl, L.; Starkl, M. Discussion on sustainable water technologies for peri-urban areas of Mexico City: Balancing urbanization and environmental conservation. Water 2012, 4, 739–758. [Google Scholar] [CrossRef] [Green Version]
- Mao, K.; Zhang, K.; Du, W.; Ali, W.; Feng, X.; Zhang, H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health 2020, 17, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Daughton, C.G. Wastewater surveillance for population-wide Covid-19: The present and future. Sci. Total Environ. 2020, 736, 139631. [Google Scholar] [CrossRef]
- Sims, N.; Kasprzyk-Hordern, B. Future perspectives of wastewater-based epidemiology: Monitoring infectious disease spread and resistance to the community level. Environ. Int. 2020, 139, 105689. [Google Scholar] [CrossRef] [PubMed]
- Pitso, L.; Potgieter, S.; Dijk, V.D.S.V.A. Prevalence of isoniazid resistance-conferring mutations associated with multidrug-resistant tuberculosis in Free State Province, South Africa. S. Afr. Med. J. 2019, 109, 659–664. [Google Scholar] [CrossRef] [Green Version]
- Tehrani, A.H.; Kimberley, A.G. A closer look at the antibiotic-resistant bacterial community found in urban wastewater treatment systems. Microbiol. Open 2018, 7, e00589. [Google Scholar] [CrossRef]
- Rodwell, C.R.; Steven, A.P.; Tomas, O.H. Patterns of age-0 yellow perch growth, diets, and mortality in Saginaw Bay, Lake Huron. J. Great Lakes Res. 2014, 40, 123–132. [Google Scholar]
- Department of Health (DOH) Republic of South Africa. Introduction of New Drugs and Drug Regimens for the Management of Drug-Resistant Tuberculosis in South Africa: Policy Framework Version; National Department of Health: Pretoria, South Africa, 2015.
- Tiberi, S.; Scardigli, A.; Centis, R.; D’Ambrosio, L.; Munoz-Torrico, M.; Salazar-Lezama, M.A.; Spanevello, A.; Visca, D.; Zumla, A.; Migliori, G.B.; et al. Classifying new anti-tuberculosis drugs: Rationale and future perspectives. Int. J. Infect. Dis. 2017, 56, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Goldstein, B.P. Resistance to rifampicin: A review. J. Antibiot. 2014, 67, 625–630. [Google Scholar] [CrossRef] [Green Version]
- Bea, S.; Lee, H.; Kim, J.H.; Jang, S.H.; Son, H.; Kwon, J.W.; Shin, J.Y. Adherence and associated factors of treatment regimen in drug-susceptible tuberculosis patients. Front. Pharmacol. 2021, 12, 625078. [Google Scholar] [CrossRef] [PubMed]
- Ektefaie, Y.; Dixit, A.; Freschi, L.; Farhat, M.R. Globally diverse Mycobacterium tuberculosis resistance acquisition: A retrospective geographical and temporal analysis of whole genome sequences. Lancet Microbe 2021, 2, e96–e104. [Google Scholar] [CrossRef]
- Kadigi, D.M.; Mosha, F.; Moyo, S.; Matee, M.I. Etiology and antimicrobial susceptibility patterns of bacterial agents causing urinary tract infection in children under five years, dar es salaam. J. Biotechnol. Immunol. 2020, 2, 2. [Google Scholar]
- Zhang, Y.; Yew, W.W. Mechanisms of drug resistance in Mycobacterium tuberculosis: Update 2015. Int. J. Tuberc. Lung Dis. 2015, 19, 1276–1289. [Google Scholar] [CrossRef] [PubMed]
- Lakshminarayana, S.B.; Huat, T.B.; Ho, P.C.; Manjunatha, U.H.; Dartois, V.; Dick, T.; Rao, S.P. Comprehensive physicochemical, pharmacokinetic and activity profiling of anti-TB agents. J. Antimicrob. Chemother. 2015, 70, 857–867. [Google Scholar] [CrossRef]
- Cuevas-Córdoba, B.; Cuellar-Sánchez, A.; Pasissi-Crivelli, A.; Santana-Álvarez, C.A.; Hernández-Illezcas, J.; Zenteno-Cuevas, R. rrs and rpsL mutations in streptomycin-resistant isolates of Mycobacterium tuberculosis from Mexico. J. Microbiol. Immunol. Infect. 2013, 46, 30–34. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Córdoba, B.; Xochihua-González, S.O.; Cuellar, A.; Fuentes-Domínguez, J.; Zenteno-Cuevas, R. Characterization of pncA gene mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from Mexico. Infect. Genet. Evol. 2013, 19, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Farhat, M.R.; Sultana, R.; Iartchouk, O.; Bozeman, S.; Galagan, J.; Sisk, P.; Stolte, C.; Nebenzahl-Guimaraes, H.; Jacobson, K.; Sloutsky, A.; et al. Genetic determinants of drug resistance in Mycobacterium tuberculosis and their diagnostic value. Am. J. Res. Crit. Care Med. 2016, 194, 621–630. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhu, X.F.; Zhou, M.G.; Kuang, J.; Zhang, Y.; Shang, Y.; Wang, J.X. Status of streptomycin resistance development in Xanthomonas oryzae pv. oryzae and Xanthomonas oryzae pv. oryzicola in China and their resistance characters. J. Phytopathol. 2010, 158, 601–608. [Google Scholar]
- Demeke, T.; Dobnik, D. Critical assessment of digital PCR for the detection and quantification of genetically modified organisms. Anal. Bioanal. Chem. 2018, 410, 4039–4050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Bai, R.; Zhao, Z.; Tao, L.; Ma, M.; Ji, Z.; Jian, M.; Ding, Z.; Dai, X.; Bao, F.; et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci. Rep. 2018, 38, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Z.; Wu, W.; Wei, H.; Gao, C.; Zhang, L.; Wu, C.; Hou, L. Using droplet digital PCR in the detection of Mycobacterium tuberculosis DNA in FFPE samples. Int. J. Infect. Dis. 2020, 99, 77–83. [Google Scholar] [CrossRef]
- Ghosh, A.; Saran, N.; Sudipto, S. Survey of drug resistance associated gene mutations in Mycobacterium tuberculosis, ESKAPE and other bacterial species. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Molin, M.D.; Gut, M.; Rominski, A.; Haldimann, K.; Becker, K.; Sander, P. Molecular mechanisms of intrinsic streptomycin resistance in Mycobacterium abscessus. Antimicrob. Agents Chemother. 2018, 62, e01427-17. [Google Scholar]
- Dai, R.; He, J.; Zha, X.; Wang, Y.; Zhang, X.; Gao, H.; Yang, X.; Li, J.; Xin, Y.; Wang, Y.; et al. A novel mechanism of streptomycin resistance in Yersinia pestis: Mutation in the rpsL gene. PLoS Negl. Trop. Dis. 2021, 15, e0009324. [Google Scholar] [CrossRef]
- Cohen, K.A.; Stott, K.E.; Munsamy, V.; Manson, A.L.; Earl, A.M.; Pym, A.S. Evidence for expanding the role of streptomycin in the management of drug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2020, 64, e00860-20. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Q.; Gao, H.; Zhang, Z.; Liu, Y.; Lu, J.; Dai, E. The roles of rpsL, rrs, and gidB mutations in predicting streptomycin-resistant drugs used on clinical Mycobacterium tuberculosis isolates from Hebei Province, China. Int. J. Clin. Exp. Pathol. 2019, 12, 2713. [Google Scholar]
- Khosravi, A.D.; Etemad, N.; Hashemzadeh, M.; Dezfuli, S.K.; Goodarzi, H. Frequency of rrs and rpsL mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from Iranian patients. J. Glob. Antimicrob. Resist. 2017, 9, 51–56. [Google Scholar] [CrossRef]
- Oo, N.A.T.; San, L.L.; Thapa, J.; Aye, K.S.; Aung, W.W.; Nakajima, C.; Suzuki, Y. Characterization of mutations conferring streptomycin resistance to multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. Tuberculosis 2018, 111, 8–13. [Google Scholar]
- Florou, Z.; Mavroidi, A.; Vatidis, G.; Daniil, Z.; Gourgoulianis, K.; Petinaki, E. Molecular Basis of Resistance to First-Line Drugs of Mycobacterium tuberculosis/canettii Strains in Greece. Microb. Drug Resist. 2021, 27, 1389–1396. [Google Scholar] [CrossRef] [PubMed]
- Padda, I.S.; Reddy, K.M. Antitubercular Medications. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557666/ (accessed on 27 October 2021).
- Kraemer, S.A.; Ramachandran, A.A.; Perron, G.G. Antibiotic pollution in the environment: From microbial ecology to public policy. Microorganisms 2019, 7, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allana, S.; Shashkina, E.; Mathema, B.; Bablishvili, N.; Tukvadze, N.; Shah, N.S.; Kempker, R.R.; Blumberg, H.M.; Moodley, P.; Mlisana, K.; et al. pncA gene mutations associated with pyrazinamide resistance in drug-resistant tuberculosis, South Africa and Georgia. Emerg. Infect. Dis. 2017, 23, 491. [Google Scholar] [CrossRef]
- Khan, M.T.; Malik, S.I.; Ali, S.; Masood, N.; Nadeem, T.; Khan, A.S.; Afzal, M.T. Pyrazinamide resistance and mutations in pncA among isolates of Mycobacterium tuberculosis from Khyber Pakhtunkhwa, Pakistan. BMC Infect. Dis. 2019, 19, 116. [Google Scholar] [CrossRef] [Green Version]
- Naluyange, R.; Mboowa, G.; Komakech, K.; Semugenze, D.; Kateete, D.P.; Ssengooba, W. High prevalence of phenotypic pyrazinamide resistance and its association with pncA gene mutations in Mycobacterium tuberculosis isolates from Uganda. PLoS ONE 2020, 15, e0232543. [Google Scholar] [CrossRef]
- Schnippel, K.; Ndjeka, N.; Maartens, G.; Meintjes, G.; Master, I.; Ismail, N.; Hughes, J.; Ferreira, H.; Padanilam, X.; Romero, R.; et al. Effect of bedaquiline on mortality in South African patients with drug-resistant tuberculosis: A retrospective cohort study. Lancet Respir. Med. 2018, 6, 699–706. [Google Scholar] [CrossRef]
- Hennebique, A.; Bidart, M.; Jarraud, S.; Beraud, L.; Schwebel, C.; Maurin, M.; Boisset, S. Digital PCR for detection and quantification of fluoroquinolone resistance in Legionella pneumophila. Antimicrob. Agents Chemother. 2017, 61, e00628-17. [Google Scholar] [CrossRef] [Green Version]
- Barancheshme, F.; Munir, M. Development of antibiotic resistance in wastewater treatment plants. In Antimicrobial Resistance-A Global Threat; IntechOpen: London, UK, 2019. [Google Scholar]
- Aali, R.; Nikaeen, M.; Khanahmad, H.; Hassanzadeh, A. Monitoring and comparison of antibiotic resistant bacteria and their resistance genes in municipal and hospital wastewaters. Int. J. Prev. Med. 2014, 5, 887. [Google Scholar] [PubMed]
- Jean-Francois, L.; Dumoutier, N. Non-tuberculous mycobacteria in drinking water systems: A review of prevalence data and control means. Int. J. Hyg. Environ. Health 2019, 222, 628–634. [Google Scholar]
- Abasiofiok, I.M.; Murinda, S.E. Linking microbial community composition in treated wastewater with water quality in distribution systems and subsequent health effects. Microorganisms 2019, 7, 660. [Google Scholar]
- Cydzik-Kwiatkowska, A.; Magdalena, Z. Bacterial communities in full-scale wastewater treatment systems. World J. Microbiol. Biotechnol. 2016, 32, 66. [Google Scholar] [CrossRef] [Green Version]
- Claeys, T.A.; Robinson, T.R. The many lives of nontuberculous mycobacteria. J. Bacteriol. 2018, 200, e00739-17. [Google Scholar] [CrossRef] [Green Version]
- Cui, B.; Liang, S. Monitoring opportunistic pathogens in domestic wastewater from a pilot-scale anaerobic biofilm reactor to reuse in agricultural irrigation. Water 2019, 11, 1283. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.S.; Kim, K.J.; Choi, H.; Lee, S.H. Delamanid, bedaquiline, and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann. Lab. Med. 2018, 38, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Samer, M. Biological and chemical wastewater treatment processes. In Wastewater Treatment Engineering; IntechOpen: London, UK, 2015. [Google Scholar]
- Jäger, T.; Hembach, N.; Elpers, C.; Wieland, A.; Alexander, J.; Hiller, C.; Krauter, G.; Schwartz, T. Reduction of antibiotic resistant bacteria during conventional and advanced wastewater treatment, and the disseminated loads released to the environment. Front. Microbiol. 2018, 9, 2599. [Google Scholar] [CrossRef] [Green Version]
- Manaia, C.M.; Macedo, G.; Fatta-Kassinos, D.; Nunes, O.C. Antibiotic resistance in urban aquatic environments: Can it be controlled? Appl. Microbiol. Biotechnol. 2016, 100, 1543–1557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzeminski, P.; Tomei, M.C.; Karaolia, P.; Langenhoff, A.; Almeida, C.M.; Felis, E.; Gritten, F.; Andersen, H.R.; Fernandes, T.; Manaia, C.M.; et al. Performance of secondary wastewater treatment methods for the removal of contaminants of emerging concern implicated in crop uptake and antibiotic resistance spread: A review. Sci. Total Environ. 2019, 648, 1052–1081. [Google Scholar] [CrossRef] [Green Version]
- Samba-Louaka, A.; Robino, E.; Cochard, T.; Branger, M.; Delafont, V.; Aucher, W.; Wambeke, W.; Bannantine, J.P.; Biet, F.; Héchard, Y. Environmental Mycobacterium avium subsp. paratuberculosis hosted by free-living amoebae. Front. Cell. Infect. Microbiol. 2018, 8, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barancheshme, F.; Munir, M. Strategies to combat antibiotic resistance in the wastewater treatment plants. Front. Microbiol. 2018, 8, 2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, S.; Cui, P.; Shi, W.; Zhang, W.; Zhang, Y. Identification of novel mutations associated with cycloserine resistance in Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2017, 72, 3272–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cross, X.; Buckley, C. SFD Promotion Initiative. SFD Report Durban, South Africa. 2016. Available online: www.sfd.susana.org. (accessed on 14 March 2021).
- Farah Aldour, M.S.; Elhussein, A.R.; Elkhidir, I.M.; Tayeib, S.E.; Mohammed Khair, O.; Mohamed, N.S.; Enan, K.A. Detection of Drug Resistant Genes of Mycobacterium Tuberculosis in Sudanese Tuberculosis Patients in Khartoum State Using Multiplex PCR. EC Microbiol. 2018, 14, 686–693. [Google Scholar]
- Pérez-Osorio, A.C.; Boyle, D.S.; Ingham, Z.K.; Ostash, A.; Gautom, R.K.; Colombel, C.; Houze, Y.; Leader, B.T. Rapid identification of mycobacteria and drug-resistant Mycobacterium tuberculosis by use of a single multiplex PCR and DNA sequencing. J. Clin. Microbiol. 2012, 50, 326–336. [Google Scholar] [CrossRef] [Green Version]
- Pillay, L.; Amoah, I.D.; Deepnarain, N.; Pillay, K.; Awolusi, O.O.; Kumari, S.; Bux, F. Monitoring changes in COVID-19 infection using wastewater-based epidemiology: A South African perspective. Sci. Total Environ. 2021, 786, 147273. [Google Scholar] [CrossRef]
- World Health Organization. Briefing Note: Antimicrobial Resistance: An Emerging Water, Sanitation and Hygiene Issue; World Health Organization: Geneva, Switzerland, 2014. [Google Scholar]




| Treatment Works | Design Capacity (Mℓ/d) | Hydraulic Retention Time (h) * | Primary Settling | Activated Sludge | Secondary Clarification | Bio-Filters | Sludge Digestion | Tertiary Treatment | Remarks |
|---|---|---|---|---|---|---|---|---|---|
| WWTP A | 18.8 | 31 | Y | N | N | Y | Y | Y | Receives from wastewater from a hospital that offers health services to the community at regional and district levels and has 17 clinics attached. This hospital is one of the sites for Mother-to-Child-Transmission (MTCT) of HIV and has the largest crisis centre. |
| WWTP B | 4.90 | 36 | N/A | Extended Aeration | Y | N | N | Y | Receives from a hospital that serves predominantly residential areas. This is also a referral hospital for another hospital and clinic |
| WWTP C | 70.0 | 38 | Y | Conventional | Y | N | Y | Y | Receives from a hospital complex which offers specialized services for Multidrug-resistant (MDR) and complicated TB |
| Chemical Class | Drug Name | Gene | References | |
|---|---|---|---|---|
| First-line drugs | Pyridine | Isoniazid (H) | katG | [11,33,34,35,36] |
| Rifamycin | Rifampicin (R) | rpoB | ||
| Ethylenediamine | Ethambutol (E) | embB | ||
| Pyrazine | Pyrazinamide (Z) | pncA | ||
| Add-on drug | Aminoglycoside | Streptomycin (S) | rrs | |
| Second-line drugs | Fluoroquinolone | Ofloxacin (Ofx) | gyrA | [11,34,37] |
| Moxifloxacin (Mfx) | gyrB | |||
| Diarylquinoline | Bedaquiline (Bdq) | atpE | ||
| Pyridine | Ethionamide (Eto) | ethR | [11,33,34] | |
| Injectable drugs | Aminoglycoside | Kanamycin (Km) | eis | [11,33,34] |
| Amikacin (Amk) | eis |
| Percentage (%) of Samples Positive (* n = 9 per Sampling Point) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Category of Treatment | Gene Detected | Antibiotic/s | WWTP A | WWTP B | WWTP C | |||||||||
| Conventional PCR | ddPCR | Conventional PCR | ddPCR | Conventional PCR | ddPCR | |||||||||
| Influent | Effluent | Influent | Effluent | Influent | Effluent | Influent | Effluent | Influent | Effluent | Influent | Effluent | |||
| First-line TB treatment | katG | H | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 100 |
| rpoB | R | 100 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| embB | E | 100 | 100 | 100 | 100 | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 100 | |
| pncA | Z | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | |
| rrs | S | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| Second-line TB treatment Other drugs | gyrA | Ofx | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 100 | 100 | 100 | 100 |
| gyrB | Mfx | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| atpE | Bdq | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| ethR | Eto | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| Injectables | eis | Km/Amk | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mtetwa, H.N.; Amoah, I.D.; Kumari, S.; Bux, F.; Reddy, P. Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa. Antibiotics 2021, 10, 1362. https://doi.org/10.3390/antibiotics10111362
Mtetwa HN, Amoah ID, Kumari S, Bux F, Reddy P. Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa. Antibiotics. 2021; 10(11):1362. https://doi.org/10.3390/antibiotics10111362
Chicago/Turabian StyleMtetwa, Hlengiwe N., Isaac D. Amoah, Sheena Kumari, Faizal Bux, and Poovendhree Reddy. 2021. "Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa" Antibiotics 10, no. 11: 1362. https://doi.org/10.3390/antibiotics10111362
APA StyleMtetwa, H. N., Amoah, I. D., Kumari, S., Bux, F., & Reddy, P. (2021). Wastewater-Based Surveillance of Antibiotic Resistance Genes Associated with Tuberculosis Treatment Regimen in KwaZulu Natal, South Africa. Antibiotics, 10(11), 1362. https://doi.org/10.3390/antibiotics10111362