Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal
Abstract
:1. Introduction
2. Materials and Methods
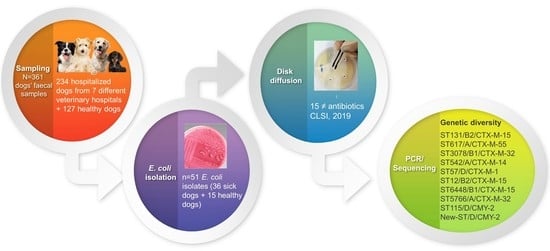
2.1. Animals and Sampling
2.2. E. coli Isolation
2.3. Susceptibility Testing
2.4. DNA Extraction and Quantification
2.5. Antibiotic Resistance and Virulence Genes Detection
2.6. Multilocus Sequence Typing and Phylogroup Typing of E. coli Isolates
2.7. Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Liu, Z.; Zhang, Y.; Zhang, Z.; Lei, L.; Xia, Z. Increasing Prevalence of ESBL-Producing Multidrug Resistance Escherichia coli From Diseased Pets in Beijing, China From 2012 to 2017. Front. Microbiol. 2019, 10, 2852. [Google Scholar] [CrossRef]
- de Melo, L.C.; Oresco, C.; Leigue, L.; Netto, H.M.; Melville, P.A.; Benites, N.R.; Saras, E.; Haenni, M.; Lincopan, N.; Madec, J.-Y. Prevalence and molecular features of ESBL/pAmpC-producing Enterobacteriaceae in healthy and diseased companion animals in Brazil. Vet. Microbiol. 2018, 221, 59–66. [Google Scholar] [CrossRef]
- Bortolami, A.; Zendri, F.; Maciuca, E.I.; Wattret, A.; Ellis, C.; Schmidt, V.; Pinchbeck, G.; Timofte, D. Diversity, Virulence, and Clinical Significance of Extended-Spectrum β-Lactamase- and pAmpC-Producing Escherichia coli From Companion Animals. Front. Microbiol. 2019, 10, 1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourne, J.A.; Chong, W.L.; Gordon, D.M. Genetic structure, antimicrobial resistance and frequency of human associated Escherichia coli sequence types among faecal isolates from healthy dogs and cats living in Canberra, Australia. PLoS ONE 2019, 14, e0212867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiot-ics. 2017. Available online: https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed on 15 January 2021).
- Dupouy, V.; Abdelli, M.; Moyano, G.; Arpaillange, N.; Bibbal, D.; Cadiergues, M.-C.; Lopez-Pulin, D.; Sayah-Jeanne, S.; De Gunzburg, J.; Saint-Lu, N.; et al. Prevalence of Beta-Lactam and Quinolone/Fluoroquinolone Resistance in Enterobacteriaceae From Dogs in France and Spain—Characterization of ESBL/pAmpC Isolates, Genes, and Conjugative Plasmids. Front. Vet. Sci. 2019, 6, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, I.; Silva, N.; Carrola, J.; Silva, V.; Currie, C.; Igrejas, G.; Poeta, P. Immunity-Acquired Resistance: Evolution of Antimicrobial Resistance Among Extended-Spectrum β-Lactamases and Carbapenemases in Klebsiella pneumoniae and Escherichia coli. In Antibiotic Drug Resistance Chapter 11; John Wiley & Sons: Hoboken, NJ, USA, 2019; pp. 239–259. [Google Scholar] [CrossRef]
- Salgado-Caxito, M.; Benavides, J.A.; Adell, A.D.; Paes, A.C.; Moreno-Switt, A.I. Global prevalence and molecular characterization of extended-spectrum β-lactamase producing-Escherichia coli in dogs and cats—A scoping review and meta-analysis. One Health 2021, 12, 100236. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Salgado-Caxito, M.; Opazo-Capurro, A.; González Muñoz, P.; Piñeiro, A.; Otto Medina, M.; Rivas, L.; Munita, J.; Millán, J. ESBL-Producing Escherichia coli Carrying CTX-M Genes Circulating among Livestock, Dogs, and Wild Mammals in Small-Scale Farms of Central Chile. Antibiotics 2021, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Bi, Z.; Ma, S.; Chen, B.; Cai, C.; He, J.; Schwarz, S.; Sun, C.; Zhou, Y.; Yin, J.; et al. Inter-host Transmission of Carbapenemase-Producing Escherichia coli among Humans and Backyard Animals. Environ. Health Perspect. 2019, 127, 107009. [Google Scholar] [CrossRef] [Green Version]
- Haenni, M.; Saras, E.; Métayer, V.; Médaille, C.; Madec, J.-Y. High Prevalence of blaCTX-M-1/IncI1/ST3 and blaCMY-2/IncI1/ST2 Plasmids in Healthy Urban Dogs in France. Antimicrob. Agents Chemother. 2014, 58, 5358–5362. [Google Scholar] [CrossRef] [Green Version]
- Belas, A.; Salazar, A.S.; Gama, L.; Couto, N.; Pomba, C. Risk factors for faecal colonisation with Escherichia coli producing extended-spectrum and plasmid-mediated AmpC β-lactamases in dogs. Vet. Rec. 2014, 175, 202. [Google Scholar] [CrossRef]
- Damborg, P.; Morsing, M.K.; Petersen, T.; Bortolaia, V.; Guardabassi, L. CTX-M-1 and CTX-M-15-producing Escherichia coli in dog faeces from public gardens. Acta Vet. Scand. 2015, 57, 83. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Sanz, E.; Torres, C.; Ceballos, S.; Lozano, C.; Zarazaga, M. Clonal Dynamics of Nasal Staphylococcus aureus and Staphylococcus pseudintermedius in Dog-Owning Household Members. Detection of MSSA ST398. PLoS ONE 2013, 8, e69337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boehmer, T.; Vogler, A.J.; Thomas, A.; Sauer, S.; Hergenroether, M.; Straubinger, R.K.; Birdsell, D.; Keim, P.; Sahl, J.W.; Williamson, C.H.D.; et al. Phenotypic characterization and whole genome analysis of extended-spectrum beta-lactamase-producing bacteria isolated from dogs in Germany. PLoS ONE 2018, 13, e0206252. [Google Scholar] [CrossRef]
- Kaspar, U.; Von Lützau, A.; Schlattmann, A.; Roesler, U.; Köck, R.; Becker, K. Zoonotic multidrug-resistant microorganisms among small companion animals in Germany. PLoS ONE 2018, 13, e0208364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, C.; Belas, A.; Franco, A.; Aboim, C.; Gama, L.; Pomba, C. Increase in antimicrobial resistance and emergence of major international high-risk clonal lineages in dogs and cats with urinary tract infection: 16 year retrospective study. J. Antimicrob. Chemother. 2017, 73, 377–384. [Google Scholar] [CrossRef] [Green Version]
- Zogg, A.L.; Simmen, S.; Zurfluh, K.; Stephan, R.; Schmitt, S.N.; Nüesch-Inderbinen, M. High Prevalence of Extended-Spectrum β-Lactamase Producing Enterobacteriaceae Among Clinical Isolates from Cats and Dogs Admitted to a Veterinary Hospital in Switzerland. Front. Vet. Sci. 2018, 5, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigg, A.; Brilhante, M.; Dazio, V.; Clément, M.; Collaud, A.; Brawand, S.G.; Willi, B.; Endimiani, A.; Schuller, S.; Perreten, V. Shedding of OXA-181 carbapenemase-producing Escherichia coli from companion animals after hospitalisation in Switzerland: An outbreak in 2018. Eurosurveillance 2019, 24, 1900071. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, J.; Schoormans, A.; Kwakernaak, M.; Duim, B.; Broens, E.; Dierikx, C.; Mevius, D.; Wagenaar, J.A. High prevalence of fecal carriage of extended spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013, 4, 242. [Google Scholar] [CrossRef] [Green Version]
- Brilhante, M.; Menezes, J.; Belas, A.; Feudi, C.; Schwarz, S.; Pomba, C.; Perreten, V. OXA-181-Producing Extraintestinal Pathogenic Escherichia coli Sequence Type 410 Isolated from a Dog in Portugal. Antimicrob. Agents Chemother. 2020, 64, e02298-19. [Google Scholar] [CrossRef]
- Costa, D.; Poeta, P.; Briñas, L.; Sáenz, Y.; Rodrigues, J.; Torres, C. Detection of CTX-M-1 and TEM-52 β-lactamases in Escherichia coli strains from healthy pets in Portugal. J. Antimicrob. Chemother. 2004, 54, 960–961. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, I.; Safia Chenouf, N.; Cunha, R.; Martins, C.; Pimenta, P.; Pereira, A.R.; Martínez-Álvarez, S.; Ramos, S.; Silva, V.; Igrejas, G.; et al. Antimicrobial Resistance Genes and Diversity of Clones among ESBL—And Acquired AmpC-Producing Escherichia coli Isolated from Fecal Samples of Healthy and Sick Cats in Portugal. Antibiotics 2021, 10, 262. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Clinical and Laboratory Standards Institute. In Performed Standards for Antimicrobial Susceptibility Testing, 29th ed.; CLSI Supplement: Wayne, PA, USA, 2019. [Google Scholar]
- Holmes, D.; Quigley, M. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 1981, 114, 193–197. [Google Scholar] [CrossRef]
- Ruiz, E.; Sáenz, Y.; Zarazaga, M.; Rocha-Gracia, R.; Martínez, L.M.; Arlet, G.; Torres, C. qnr, aac(6′)-Ib-cr and qepA genes in Escherichia coli and Klebsiella spp.: Genetic environments and plasmid and chromosomal location. J. Antimicrob. Chemother. 2012, 67, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Zong, Z.; Partridge, S.R.; Thomas, L.; Iredell, J.R. Dominance of blaCTX-M within an Australian Extended-Spectrum β-Lactamase Gene Pool. Antimicrob. Agents Chemother. 2008, 52, 4198–4202. [Google Scholar] [CrossRef] [Green Version]
- Pitout, J.D.; Thomson, K.S.; Hanson, N.D.; Ehrhardt, A.F.; Moland, E.S.; Sanders, C.C. β-Lactamases responsible for resistance to expanded-spectrum cephalosporins in Klebsiella pneumoniae, Escherichia coli, and Proteus mirabilis isolates recovered in South Africa. Antimicrob. Agents Chemother. 1998, 42, 1350–1354. [Google Scholar] [CrossRef] [Green Version]
- Porres-Osante, N.; Azcona-Gutiérrez, J.M.; Rojo-Bezares, B.; Undabeitia, E.; Torres, C.; Sáenz, Y. Emergence of a multiresistant KPC-3 and VIM-1 carbapenemase-producing Escherichia coli strain in Spain. J. Antimicrob. Chemother. 2014, 69, 1792–1795. [Google Scholar] [CrossRef] [Green Version]
- Vinué, L.; Sáenz, Y.; Somalo, S.; Escudero, E.; Moreno, M.A.; Ruiz-Larrea, F.; Torres, C. Prevalence and diversity of integrons and associated resistance genes in faecal Escherichia coli isolates of healthy humans in Spain. J. Antimicrob. Chemother. 2008, 62, 934–937. [Google Scholar] [CrossRef] [Green Version]
- Hassen, B.; Abbassi, M.S.; Ruiz-Ripa, L.; Mama, O.M.; Hassen, A.; Torres, C.; Hammami, S. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int. J. Food Microbiol. 2019, 318, 108478. [Google Scholar] [CrossRef]
- Ellington, M.J.; Kistler, J.J.; Livermore, D.M.; Woodford, N. Multiplex PCR for rapid detection of genes encoding acquired metallo-lactamases. J. Antimicrob. Chemother. 2006, 59, 321–322. [Google Scholar] [CrossRef] [Green Version]
- Vidal, R.; Vidal, M.; Lagos, R.; Levine, M.; Prado, V. Multiplex PCR for Diagnosis of Enteric Infections Associated with Diarrheagenic Escherichia coli. J. Clin. Microbiol. 2004, 42, 1787–1789. [Google Scholar] [CrossRef] [Green Version]
- PubMLST. Escherichia coli (Achtman) MLST Locus/Sequence Definitions Database. Available online: https://pubmlst.org/bigsdb?db=pubmlst_ecoli_achtman_seqdef (accessed on 15 January 2021).
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.; González-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carvalho, A.; Barbosa, A.; Arais, L.; Ribeiro, P.; Carneiro, V.; Cerqueira, A. Resistance patterns, ESBL genes, and genetic relatedness of Escherichia coli from dogs and owners. Braz. J. Microbiol. 2016, 47, 150–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Zeng, Z.; Chen, S.; Ma, J.; He, L.; Liu, Y.; Deng, Y.; Lei, T.; Zhao, J.; Liu, J.-H. High prevalence of blaCTX-M extended-spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin. Microbiol. Infect. 2010, 16, 1475–1481. [Google Scholar] [CrossRef] [PubMed]
- Ben Sallem, R.; Gharsa, H.; BEN Slama, K.; Rojo-Bezares, B.; Estepa, V.; Porres-Osante, N.; Jouini, A.; Klibi, N.; Sáenz, Y.; Boudabous, A.; et al. First Detection of CTX-M-1, CMY-2, and QnrB19 Resistance Mechanisms in Fecal Escherichia coli Isolates from Healthy Pets in Tunisia. Vector Borne Zoonotic Dis. 2013, 13, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Dahmen, S.; Haenni, M.; Châtre, P.; Madec, J.-Y. Characterization of blaCTX-M IncFII plasmids and clones of Escherichia coli from pets in France. J. Antimicrob. Chemother. 2013, 68, 2797–2801. [Google Scholar] [CrossRef] [Green Version]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic Resistance and Virulence Traits in Vancomycin-Resistant Enterococci (VRE) and Extended-Spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from Humans and Pets. Antibiotics 2020, 9, 152. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Navarro, J.; Miro, E.; Brown-Jaque, M.; Hurtado, J.C.; Moreno, A.; Muniesa, M.; Gonzalez-Lopez, J.J.; Vila, J.; Espinal, P.; Navarro, F. Diversity of plasmids in Escherichia coli and Klebsiella pneumoniae: A comparison of commensal and clinical isolates. Antimicrob. Agents Chemother. 2020, 64. [Google Scholar] [CrossRef]
- Kawamura, K.; Sugawara, T.; Matsuo, N.; Hayashi, K.; Norizuki, C.; Tamai, K.; Kondo, T.; Arakawa, Y. Spread of CTX-Type Extended-Spectrum β-Lactamase-Producing Escherichia coli Isolates of Epidemic Clone B2-O25-ST131 Among Dogs and Cats in Japan. Microb. Drug Resist. 2017, 23, 1059–1066. [Google Scholar] [CrossRef]
- Aslantaş, O.; Yilmaz, E. Prevalence and molecular characterization of extended-spectrum β-lactamase (ESBL) and plasmidic AmpC β-lactamase (pAmpC) producing Escherichia coli in dogs. J. Vet. Med Sci. 2017, 79, 1024–1030. [Google Scholar] [CrossRef] [Green Version]
- Costa, D.; Poeta, P.; Sáenz, Y.; Coelho, A.C.; Matos, M.; Vinue, L.; Rodrigues, J.; Torres, C. Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol. 2008, 127, 97–105. [Google Scholar] [CrossRef]
- Meireles, D.; Leite-Martins, L.; Bessa, L.; Cunha, S.; Fernandes, R.; De Matos, A.; Manaia, C.; Da Costa, P.M. Molecular characterization of quinolone resistance mechanisms and extended-spectrum β-lactamase production in Escherichia coli isolated from dogs. Comp. Immunol. Microbiol. Infect. Dis. 2015, 41, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, M.; Irrgang, A.; Roschanski, N.; Michael, G.B.; Hamprecht, A.; Rieber, H.; Käsbohrer, A.; Schwarz, S.; Rösler, U. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genom. 2018, 19, 601. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Betts, J.; La Ragione, R.; van Essen-Zandbergen, A.; Ceccarelli, D.; Petinaki, E.; Koutinas, C.K.; Mevius, D.J. Occurrence and characterization of extended-spectrum cephalosporin-resistant Enterobacteriaceae in healthy household dogs in Greece. J. Med. Microbiol. 2018, 67, 931–935. [Google Scholar] [CrossRef]
- Belas, A.; Marques, C.; Aboim, C.; Pomba, C. Emergence of Escherichia coli ST131 H30/H30-Rx subclones in companion animals. J. Antimicrob. Chemother. 2018, 74, 266–269. [Google Scholar] [CrossRef] [Green Version]
- Rocha-Gracia, R.; Cortés-Cortés, G.; Lozano-Zarain, P.; Bello, F.; Martínez-Laguna, Y.; Torres, C. Faecal Escherichia coli isolates from healthy dogs harbour CTX-M-15 and CMY-2 β-lactamases. Vet. J. 2014, 203, 315–319. [Google Scholar] [CrossRef]
- Carvalho, I.; Tejedor-Junco, M.T.; González-Martín, M.; Corbera, J.A.; Suárez-Pérez, A.; Silva, V.; Igrejas, G.; Torres, C.; Poeta, P. Molecular diversity of Extended-spectrum β-lactamase-producing Escherichia coli from vultures in Canary Islands. Environ. Microbiol. Rep. 2020, 12, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Kaarme, J.; Riedel, H.; Schaal, W.; Yin, H.; Nevéus, T.; Melhus, Å. Rapid Increase in Carriage Rates of Enterobacteriaceae Producing Extended-Spectrum β-Lactamases in Healthy Preschool Children, Sweden. Emerg. Infect. Dis. 2018, 24, 1874–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birgy, A.; Madhi, F.; Jung, C.; Levy, C.; Cointe, A.; Bidet, P.; Hobson, C.A.; Bechet, S.; Sobral, E.; Vuthien, H.; et al. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J. Antimicrob. Chemother. 2019, 75, 96–105. [Google Scholar] [CrossRef]
- Mhaya, A.; Trabelsi, R.; Begu, D.; Aillerie, S.; M’zali, F.; Tounsi, S.; Gdoura, R.; Arpin, C. Emergence of B2-ST131-C2 and A-ST617 Escherichia coli clones producing both CTX-M-15 and CTX-M-27 and ST147 NDM-1 positive Klebsiella pneumoniae in the Tunisian community. bioRxiv 2019, 713461. [Google Scholar] [CrossRef]
- Ben Sallem, R.; Ben Slama, K.; Estepa, V.; Cheikhna, E.O.; Mohamed, A.M.; Chairat, S.; Ruiz-Larrea, F.; Boudabous, A.; Torres, C. Detection of CTX-M-15-producing Escherichia coli isolates of lineages ST410-A, ST617-A and ST354-D in faecal samples of hospitalized patients in a Mauritanian hospital. J. Chemother. 2014, 27, 114–116. [Google Scholar] [CrossRef]
- Gauthier, L.; Dortet, L.; Cotellon, G.; Creton, E.; Cuzon, G.; Ponties, V.; Bonnin, R.A.; Naas, T. Diversity of Carbapenemase-Producing Escherichia coli Isolates in France in 2012–2013. Antimicrob. Agents Chemother. 2018, 62, e00266-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dahms, C.; Hübner, N.-O.; Kossow, A.; Mellmann, A.; Dittmann, K.; Kramer, A. Occurrence of ESBL-Producing Escherichia coli in Livestock and Farm Workers in Mecklenburg-Western Pomerania, Germany. PLoS ONE 2015, 10, e0143326. [Google Scholar] [CrossRef] [PubMed]
- Reid, C.; Wyrsch, E.R.; Chowdhury, P.R.; Zingali, T.; Liu, M.; Darling, A.; Chapman, T.; Djordjevic, S.P. Porcine commensal Escherichia coli: A reservoir for class 1 integrons associated with IS26. Microb. Genom. 2017, 3, e000143. [Google Scholar] [CrossRef] [PubMed]
- Braga, J.F.V.; Chanteloup, N.K.; Trotereau, A.; Baucheron, S.; Guabiraba, R.; Ecco, R.; Schouler, C. Diversity of Escherichia coli strains involved in vertebral osteomyelitis and arthritis in broilers in Brazil. BMC Vet. Res. 2016, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bréchet, C.; Plantin, J.; Thouverez, M.; Cholley, P.; Bertrand, X.; Hocquet, D. The wastewater network likely plays a role in the spread of ESBL-producing Escherichia coli in the community. In Proceedings of the 23rd European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Berlin, Germany, 12 February 1983. [Google Scholar]


| Isolate Number | Origin a | Sick/Healthy | Gender b | Age c | Breed d | Phenotype of Antibiotic Resistance e | β-Lactamases | Other Genes and Integrons f | PG g | MLST h |
|---|---|---|---|---|---|---|---|---|---|---|
| X605 | HV Lisboa | Sick | M | 15A | UD | AMP, CTX, ATM, CHL, CIP, TET | CTX-M-15 | tet(A) | B1 | ST6448 |
| X614 | HV Lisboa | Sick | F | 2A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, TET | CTX-M-15, CMY-2 | tet(A) | B1 | ST6448 |
| X607 | HV Lisboa | Sick | F | 1A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST131 |
| X610 | HV Lisboa | Sick | F | 1,5A | UD | AMP, CTX, CAZ, ATM, CIP, S, TET | CTX-M-15 | tet(A) | B2 | ST12 |
| X603 | CV Bragança | Sick | F | 10A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST131 |
| X602 | CV VR | Sick | F | 3A | UD | AMP, AUG, CTX, CHL, TOB, CN, S, TET | CTX-M-15 | tet(A) | B2 | ST12 |
| X558 | Kennel | Healthy | M | 2A | Labrador | AMP, AUG, CTX, CAZ, CIP, SXT, S, TET | CTX-M-15, TEM | int1 | B1 | NT |
| X562 | Kennel | Healthy | F | 4M | UD | AMP, CTX, CIP, SXT, S, TET | CTX-M-15, TEM | tet(A), int1 | B1 | NT |
| X569 | HVTM | Sick | M | 4A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-15, TEM | tet(A), tet(B), int1 | A | NT |
| X575 | CV Transm | Sick | M | 4A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, TET | CTX-M-15, TEM | tet(A), tet(B), int1 | A | NT |
| C10151 | HV Lisboa | Sick | F | 11A | UD | AMP, AUG, CTX, TET | CTX-M-15, TEM | ND | A | NT |
| X550 | HD | Healthy | F | 8A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X556 | HD | Healthy | F | 14A | Yorkshire | AMP, AUG, FOX, CTX, ATM | CTX-M-15 | ND | B1 | NT |
| X563 | Kennel | Healthy | M | 5A | Rottweiler | AMP, AUG, CTX, TET | CTX-M-15 | ND | A | NT |
| X588 | HVTM | Sick | M | 5A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CN, TET | CTX-M-15 | tet(A) | D | NT |
| X598 | HVTM | Sick | M | 6A | Russell Terrier | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | ND | B1 | NT |
| X576 | HV Lisboa | Sick | M | 15A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | D | NT |
| X577 | HV Lisboa | Sick | F | 6M | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X578 | HV Lisboa | Sick | M | 13A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, SXT, TOB, TET | CTX-M-15 | tet(A) | B1 | NT |
| X580 | HV Lisboa | Sick | F | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | int1 | B1 | NT |
| X584 | HV Lisboa | Sick | M | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X604 | HV Lisboa | Sick | M | 12A | UD | AMP, AUG, CTX, ATM | CTX-M-15 | ND | D | NT |
| X618 | HV Lisboa | Sick | F | 2A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-15 | tet(A), int1 | B1 | NT |
| X620 | HV Lisboa | Sick | M | 9A | UD | AMP, AUG, CTX, CAZ, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-15 | tet(A), int1 | D | NT |
| X622 | HV Lisboa | Sick | M | 3A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, CIP, SXT, S, TET | CTX-M-15 | ND | A | NT |
| X599 | CV Bragança | Sick | M | 7A | Rodengo | AMP, AUG, FOX, CTX, CAZ, ATM, CHL, CIP, TET | CTX-M-15 | ND | B1 | NT |
| C10264 | CV Vouga | Sick | F | 9M | Pincher | AMP, CTX, CAZ | CTX-M-1 | ND | D | ST57 |
| X554 | HVTM | Sick | M | 1A | Labrador | AMP, CTX, CAZ, TET | CTX-M-1, TEM | ND | A | NT |
| X557 | Kennel | Healthy | F | 3A | Serra Estrela | AMP, AUG, CTX, CAZ, TOB, AK, S | CTX-M-1 | ND | B1 | NT |
| X559 | Kennel | Healthy | F | 3M | UD | AMP, AUG, CTX, CAZ | CTX-M-1 | ND | D | NT |
| X560 | Kennel | Healthy | M | 1A | Labrador | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | ND | B1 | NT |
| X581 | HV Lisboa | Sick | M | 14A | UD | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | tet(A) | B1 | NT |
| X611 | HV Lisboa | Sick | M | 5A | UD | AMP, CTX, CAZ, ATM, CHL, CIP, CN, TET | CTX-M-1 | tet(A) | D | NT |
| X616 | HV Lisboa | Sick | M | 3M | UD | AMP, AUG, CTX, CAZ, TET | CTX-M-1 | ND | B1 | NT |
| X617 | HV Lisboa | Sick | F | 3A | UD | AMP, CTX, CAZ, TET | CTX-M-1 | tet(A) | B1 | NT |
| C10265 | CV Bragança | Sick | M | 1A | UD | AMP, CTX, CAZ, S | CTX-M-1 | ND | A | NT |
| X555 | HD | Healthy | M | 6A | Pastor alemão | AMP, CTX, CAZ, ATM, CIP, SXT, TOB, CN, TET | CTX-M-55 | tet(B), int1 | A | ST617 |
| X568 | HD | Healthy | M | 7A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, CIP, SXT, TET | CTX-M-55 | tet(A), int1 | B1 | NT |
| C10149 | HVTM | Sick | F | 1,5A | UD | AMP, CTX, CAZ, CHL, TOB, CN, S, TET | CTX-M-55, TEM | tet(A) | A | NT |
| X573 | HD | Healthy | M | 1A | UD | AMP, AUG, CTX, CAZ, ATM, CHL, SXT, S, TET | CTX-M-32 | int1 | A | ST5766 |
| X561 | Kennel | Healthy | M | 2A | Gado transm. | AMP, AUG, CTX, ATM, CHL, CIP, SXT, TOB, CN, S, TET | CTX-M-32 | tet(A), int1 | B1 | ST3078 |
| X571 | HD | Healthy | M | 1A | UD | AMP, AUG, CTX, CAZ, CHL, SXT, S, TET | CTX-M-32, TEM | tet(B), int1 | B1 | NT |
| X572 | HD | Healthy | F | 1A | UD | AMP, AUG, CTX, CAZ, S, TET | CTX-M-14 | tet(B) | A | ST542 |
| X574 | CVTransm | Sick | M | 4A | UD | AMP, CTX, CHL, SXT, TOB, CN, S, TET | CTX-M-14 | ND | A | NT |
| X565 | HD | Healthy | M | 6A | UD | AMP, CTX, ATM, CIP, SXT, TOB, CN, S, TET | CTX-M-variant | ND | A | NT |
| C10147 | HVLisboa | Sick | M | 7A | UD | AMP, CTX, CHL, SXT, CN, S, TET | TEM-1 | tet(A), int1 | B2 | NT |
| X587 | HVTM | Sick | M | 2A | Bulldog Francês | AMP, CTX, ATM, CHL, SXT, CN, S, TET | No bla genes | int1 | A | NT |
| Isolate Number | Origin a | Gender b | Age c | Breed d | Antimicrobial Resistance Phenotype e | Resistance Genotype | Other Resistance Genes f | PG g | MLST h |
|---|---|---|---|---|---|---|---|---|---|
| X551 | HD | F | 24M | Golden Retriever | AMP, CTX | CMY-2 | ND | D | New ST * |
| X567 | CV Vouga | F | 8A | UD | AMP, AUG, FOX, CTX, CAZ, CIP, S, TET | CMY-2, TEM | tet(A) | D | ST115 |
| X549 | HVTM | F | 6A | Leão Rodesea | AMP, AUG, CTX | ND | ND | D | NT |
| C10266 | HV Lisboa | F | 6A | UD | AMP, AUG, FOX, CTX, CAZ, ATM, NA, CIP, SXT, S, TET | ND | tet(B) | A | NT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, I.; Cunha, R.; Martins, C.; Martínez-Álvarez, S.; Safia Chenouf, N.; Pimenta, P.; Pereira, A.R.; Ramos, S.; Sadi, M.; Martins, Â.; et al. Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics 2021, 10, 1013. https://doi.org/10.3390/antibiotics10081013
Carvalho I, Cunha R, Martins C, Martínez-Álvarez S, Safia Chenouf N, Pimenta P, Pereira AR, Ramos S, Sadi M, Martins Â, et al. Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics. 2021; 10(8):1013. https://doi.org/10.3390/antibiotics10081013
Chicago/Turabian StyleCarvalho, Isabel, Rita Cunha, Carla Martins, Sandra Martínez-Álvarez, Nadia Safia Chenouf, Paulo Pimenta, Ana Raquel Pereira, Sónia Ramos, Madjid Sadi, Ângela Martins, and et al. 2021. "Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal" Antibiotics 10, no. 8: 1013. https://doi.org/10.3390/antibiotics10081013
APA StyleCarvalho, I., Cunha, R., Martins, C., Martínez-Álvarez, S., Safia Chenouf, N., Pimenta, P., Pereira, A. R., Ramos, S., Sadi, M., Martins, Â., Façanha, J., Rabbi, F., Capita, R., Alonso-Calleja, C., de Lurdes Nunes Enes Dapkevicius, M., Igrejas, G., Torres, C., & Poeta, P. (2021). Antimicrobial Resistance Genes and Diversity of Clones among Faecal ESBL-Producing Escherichia coli Isolated from Healthy and Sick Dogs Living in Portugal. Antibiotics, 10(8), 1013. https://doi.org/10.3390/antibiotics10081013