Effects of Ciprofloxacin Alone or in Mixture with Sulfamethoxazole on the Efficiency of Anaerobic Digestion and Its Microbial Community
Abstract
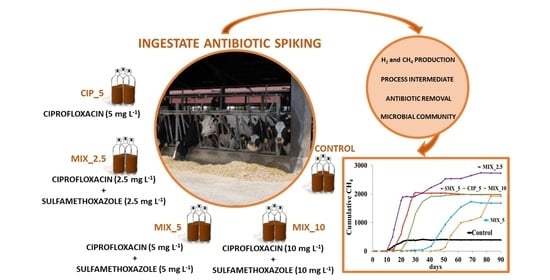
:1. Introduction
2. Results and Discussion
2.1. Biogas Production and Process Intermediates
2.2. Antibiotic Removal
2.3. Microbial Community Analysis
3. Materials and Methods
3.1. Anaerobic Digestion Test
3.2. Biogas and Organic Acid Measurements
3.3. Antibiotic Determination
3.4. Microbiological Analysis
3.5. DNA Extraction and Next Generation Sequence (NGS)
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, L.; Wang, Y.; Lichtfouse, E.; Li, Z.; Kumar, P.S.; Liu, J.; Feng, D.; Yang, Q.; Liu, F. Effect of Antibiotics on the Microbial Efficiency of Anaerobic Digestion of Wastewater: A Review. Front. Microbiol. 2021, 11, 611613. [Google Scholar] [CrossRef] [PubMed]
- Barra Caracciolo, A.; Visca, A.; Massini, G.; Patrolecco, L.; Miritana, V.M.; Grenni, P. Environmental Fate of Antibiotics and Resistance Genes in Livestock Waste and Digestate from Biogas Plants. Environ. Sci. Pollut. Res. Manag. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Talavera-Caro, A.G.; Lira, I.O.H.-D.; Cruz, E.R.; Sánchez-Muñoz, M.A.; Balagurusamy, N. The Realm of Microorganisms in Biogas Production: Microbial Diversity, Functional Role, Community Interactions, and Monitoring the Status of Biogas Plant. In Biogas Production; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Agneessens, L.M.; Ottosen, L.D.M.; Voigt, N.V.; Nielsen, J.L.; de Jonge, N.; Fischer, C.H.; Kofoed, M.V.W. In-situ biogas upgrading with pulse H2 additions: The relevance of methanogen adaption and inorganic carbon level. Bioresour. Technol. 2017, 233, 256–263. [Google Scholar] [CrossRef]
- Lehmann, L.; Bloem, E. Antibiotic residues in substrates and output materials from biogas plants—Implications for agriculture. Chemosphere 2021, 278, 130425. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, V.; Collins, G.; Mahony, T. Anaerobic Digestion of Agricultural Residues. In Environmental Microbiology; Society for Applied Microbiology and John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2010; pp. 259–279. ISBN 9780470495117. [Google Scholar]
- Hammer, T.J.; Fierer, N.; Hardwick, B.; Simojoki, A.; Slade, E.; Taponen, J.; Viljanen, H.; Roslin, T. Treating cattle with antibiotics affects greenhouse gas emissions, and microbiota in dung and dung beetles. Proc. R. Soc. B Biol. Sci. 2016, 283, 20160150. [Google Scholar] [CrossRef]
- Grenni, P. Antimicrobial Resistance in Rivers: A Review of the Genes Detected and New Challenges. Environ. Toxicol. Chem. 2022, 41, 687–714. [Google Scholar] [CrossRef]
- Cheng, D.L.; Ngo, H.H.; Guo, W.S.; Chang, S.W.; Nguyen, D.D.; Kumar, S.M.; Du, B.; Wei, Q.; Wei, D. Problematic effects of antibiotics on anaerobic treatment of swine wastewater. Bioresour. Technol. 2018, 263, 642–653. [Google Scholar] [CrossRef]
- Kumar, K.; Gupta, S.C.; Chander, Y.; Singh, A.K. Antibiotic Use in Agriculture and Its Impact on the Terrestrial Environment. Adv. Agron. 2005, 87, 1–54. [Google Scholar]
- Massé, D.I.; Saady, N.M.C.; Gilbert, Y. Potential of Biological Processes to Eliminate Antibiotics in Livestock Manure: An Overview. Animals 2014, 4, 146–163. [Google Scholar] [CrossRef]
- Rauseo, J.; Barra Caracciolo, A.; Ademollo, N.; Cardoni, M.; Di Lenola, M.; Gaze, W.H.; Stanton, I.C.; Grenni, P.; Pescatore, T.; Spataro, F.; et al. Dissipation of the antibiotic sulfamethoxazole in a soil amended with anaerobically digested cattle manure. J. Hazard. Mater. 2019, 378, 120769. [Google Scholar] [CrossRef]
- Huygens, J.; Daeseleire, E.; Mahillon, J.; Van Elst, D.; Decrop, J.; Meirlaen, J.; Dewulf, J.; Heyndrickx, M.; Rasschaert, G. Presence of antibiotic residues and antibiotic resistant bacteria in cattle manure intended for fertilization of agricultural fields: A one health perspective. Antibiotics 2021, 10, 410. [Google Scholar] [CrossRef] [PubMed]
- Agga, G.E.; Kasumba, J.; Loughrin, J.H.; Conte, E.D. Anaerobic Digestion of Tetracycline Spiked Livestock Manure and Poultry Litter Increased the Abundances of Antibiotic and Heavy Metal Resistance Genes. Front. Microbiol. 2020, 11, 614424. [Google Scholar] [CrossRef]
- Andriamalala, A.; Vieublé-Gonod, L.; Dumeny, V.; Cambier, P. Fate of sulfamethoxazole, its main metabolite N-ac-sulfamethoxazole and ciprofloxacin in agricultural soils amended or not by organic waste products. Chemosphere 2018, 191, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Grenni, P.; Ancona, V.; Barra Caracciolo, A. Ecological effects of antibiotics on natural ecosystems: A review. Microchem. J. 2018, 136, 25–39. [Google Scholar] [CrossRef]
- Visca, A.; Barra Caracciolo, A.; Grenni, P.; Patrolecco, L.; Rauseo, J.; Massini, G.; Mazzurco Miritana, V.; Spataro, F. Anaerobic Digestion and Removal of Sulfamethoxazole, Enrofloxacin, Ciprofloxacin and Their Antibiotic Resistance Genes in a Full-Scale Biogas Plant. Antibiotics 2021, 10, 502. [Google Scholar] [CrossRef]
- Girardi, C.; Greve, J.; Lamshöft, M.; Fetzer, I.; Miltner, A.; Schäffer, A.; Kästner, M. Biodegradation of ciprofloxacin in water and soil and its effects on the microbial communities. J. Hazard. Mater. 2011, 198, 22–30. [Google Scholar] [CrossRef]
- Patrolecco, L.; Rauseo, J.; Ademollo, N.; Grenni, P.; Cardoni, M.; Levantesi, C.; Luprano, M.L.; Caracciolo, A.B. Persistence of the antibiotic sulfamethoxazole in river water alone or in the co-presence of ciprofloxacin. Sci. Total Environ. 2018, 640–641, 1438–1446. [Google Scholar] [CrossRef]
- Pallo-Zimmerman, L.M.; Byron, J.K.; Graves, T.K. Fluoroquinolones: Then and now. Compend. Contin. Educ. Vet. 2010, 32, E1–E9. [Google Scholar]
- Rusch, M.; Spielmeyer, A.; Zorn, H.; Hamscher, G. Degradation and transformation of fluoroquinolones by microorganisms with special emphasis on ciprofloxacin. Appl. Microbiol. Biotechnol. 2019, 103, 6933–6948. [Google Scholar] [CrossRef]
- Müller, E.; Schüssler, W.; Horn, H.; Lemmer, H. Aerobic biodegradation of the sulfonamide antibiotic sulfamethoxazole by activated sludge applied as co-substrate and sole carbon and nitrogen source. Chemosphere 2013, 92, 969–978. [Google Scholar] [CrossRef]
- Vila-Costa, M.; Gioia, R.; Aceña, J.; Pérez, S.; Casamayor, E.O.; Dachs, J. Degradation of sulfonamides as a microbial resistance mechanism. Water Res. 2017, 115, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Cetecioglu, Z.; Ince, B.; Orhon, D.; Ince, O. Anaerobic sulfamethoxazole degradation is driven by homoacetogenesis coupled with hydrogenotrophic methanogenesis. Water Res. 2016, 90, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, B.; Zou, R.; Dai, Y.; Xie, S.; Yuan, B. Biodegradation of antibiotic ciprofloxacin: Pathways, influential factors, and bacterial community structure. Environ. Sci. Pollut. Res. 2016, 23, 7911–7918. [Google Scholar] [CrossRef]
- Silva, A.R.; Gomes, J.C.; Salvador, A.F.; Martins, G.; Alves, M.M.; Pereira, L. Ciprofloxacin, diclofenac, ibuprofen and 17α-ethinylestradiol differentially affect the activity of acetogens and methanogens in anaerobic communities. Ecotoxicology 2020, 29, 866–875. [Google Scholar] [CrossRef]
- Zhi, S.; Zhang, K. Antibiotic residues may stimulate or suppress methane yield and microbial activity during high-solid anaerobic digestion. Chem. Eng. J. 2019, 359, 1303–1315. [Google Scholar] [CrossRef]
- Jia, Y.; Khanal, S.K.; Shu, H.; Zhang, H.; Chen, G.H.; Lu, H. Ciprofloxacin degradation in anaerobic sulfate-reducing bacteria (SRB) sludge system: Mechanism and pathways. Water Res. 2018, 136, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Nguyen, L.N.; Oh, S. Ecological impact of the antibiotic ciprofloxacin on microbial community of aerobic activated sludge. Environ. Geochem. Health 2020, 42, 1531–1541. [Google Scholar] [CrossRef]
- Aydin, S.; Ince, B.; Cetecioglu, Z.; Arikan, O.; Ozbayram, E.G.; Shahi, A.; Ince, O. Combined effect of erythromycin, tetracycline and sulfamethoxazole on performance of anaerobic sequencing batch reactors. Bioresour. Technol. 2015, 186, 207–214. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Arikan, O.; Ince, B.; Cetecioglu, Z.; Aydin, S.; Ince, O. Acute effects of various antibiotic combinations on acetoclastic methanogenic activity. Environ. Sci. Pollut. Res. 2015, 22, 6230–6235. [Google Scholar] [CrossRef]
- Zhang, J.; Qi, C.; Wang, Y.; Li, Y.; Han, T.; Gong, X.; Shan, M.; Li, G.; Luo, W. Enhancing biogas production from livestock manure in solid-state anaerobic digestion by sorghum-vinegar residues. Environ. Technol. Innov. 2022, 26, 102276. [Google Scholar] [CrossRef]
- Mazzurco Miritana, V.; Massini, G.; Visca, A.; Grenni, P.; Patrolecco, L.; Spataro, F.; Rauseo, J.; Garbini, G.L.; Signorini, A.; Rosa, S.; et al. Effects of Sulfamethoxazole on the Microbial Community Dynamics during the Anaerobic Digestion Process. Front. Microbiol. 2020, 11, 537783. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Li, X.; Zilles, J.L.; Morgenroth, E.; Raskin, L. Effects of the antimicrobial tylosin on the microbial community structure of an anaerobic sequencing batch reactor. Biotechnol. Bioeng. 2011, 108, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Koniuszewska, I.; Harnisz, M.; Korzeniewska, E.; Czatzkowska, M.; Jastrzȩbski, J.P.; Paukszto, L.; Bajkacz, S.; Felis, E.; Rusanowska, P. The effect of antibiotics on mesophilic anaerobic digestion process of cattle manure. Energies 2021, 14, 1125. [Google Scholar] [CrossRef]
- Ferraro, A.; Dottorini, G.; Massini, G.; Mazzurco Miritana, V.; Signorini, A.; Lembo, G.; Fabbricino, M. Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour. Technol. 2018, 260, 364–373. [Google Scholar] [CrossRef]
- Deng, Y.; Zhang, Y.; Gao, Y.; Li, D.; Liu, R.; Liu, M.; Zhang, H.; Hu, B.; Yu, T.; Yang, M. Microbial community compositional analysis for series reactors treating high level antibiotic wastewater. Environ. Sci. Technol. 2012, 46, 795–801. [Google Scholar] [CrossRef]
- Khelaifia, S.; Drancourt, M. Susceptibility of archaea to antimicrobial agents: Applications to clinical microbiology. Clin. Microbiol. Infect. 2012, 18, 841–848. [Google Scholar] [CrossRef]
- Mustapha, N.A.; Hu, A.; Yu, C.P.; Sharuddin, S.S.; Ramli, N.; Shirai, Y.; Maeda, T. Seeking key microorganisms for enhancing methane production in anaerobic digestion of waste sewage sludge. Appl. Microbiol. Biotechnol. 2018, 102, 5323–5334. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Methanosphaera stadtmaniae gen. nov., sp. nov.: A species that forms methane by reducing methanol with hydrogen. Arch. Microbiol. 1985, 141, 116–122. [Google Scholar] [CrossRef]
- Martínez-Sibaja, A.; Alvarado-Lassman, A.; Astorga-Zaragoza, C.M.; Adam-Medina, M.; Posada-Gómez, R.; Rodríguez-Jarquin, J.P. Volumetric gas meter for laboratory-scale anaerobic bioreactors. Measurement 2011, 44, 1801–1805. [Google Scholar] [CrossRef]
- Logan, B.E.; Oh, S.E.; Kim, I.S.; Van Ginkel, S. Biological hydrogen production measured in batch anaerobic respirometers. Environ. Sci. Technol. 2002, 36, 2530–2535. [Google Scholar] [CrossRef]
- Thompson, M.; Ellison, S.L.R.; Wood, R. Laboratory validation of methods—harmonized guidelines. Pure Appl. Chem. 2002, 74, 835–855. [Google Scholar] [CrossRef]
- Pernthaler, J.; Glöckner, F.-O.; Schönhuber, W.; Amann, R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. In Marine Microbiology; Paul, J.H., Ed.; Elsevier: Amsterdam, The Netherlands, 2001; pp. 207–226. [Google Scholar]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Barra Caracciolo, A.; Grenni, P.; Cupo, C.; Rossetti, S. In situ analysis of native microbial communities in complex samples with high particulate loads. FEMS Microbiol. Lett. 2005, 253, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Barra Caracciolo, A.; Giuliano, G.; Grenni, P.; Cremisini, C.; Ciccoli, R.; Ubaldi, C. Effect of urea on degradation of terbuthylazine in soil. Environ. Toxicol. Chem. 2005, 24, 1035–1040. [Google Scholar] [CrossRef]
- Greuter, D.; Loy, A.; Horn, M.; Rattei, T. ProbeBase-an online resource for rRNA-targeted oligonucleotide probes and primers: New features 2016. Nucleic Acids Res. 2016, 44, D586–D589. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Chao, A.; Chazdon, R.L.; Shen, T.J. A new statistical approach for assessing similarity of species composition with incidence and abundance data. Ecol. Lett. 2005, 8, 148–159. [Google Scholar] [CrossRef]
- Benavoli, A.; Corani, G.; Mangili, F. Should we really use post-hoc tests based on mean-ranks? J. Mach. Learn. Res. 2016, 17, 1–10. [Google Scholar]






| Bacteria/Archaea | |||||
|---|---|---|---|---|---|
| Time (Day) | Experimental Conditions | ||||
| Control | CIP_5 | Mix_2.5 | Mix_5 | Mix_10 | |
| 3 | 18.5 | 57.7 | 27.0 | 63.6 | 66.4 |
| 6 | 19.4 | 36.9 | 33.8 | 60.8 | 141.4 |
| 10 | 18.4 | 27.6 | 70.6 | 39.6 | 102.2 |
| 15 | 30.0 | 31.0 | 30.7 | 86.4 | 133.7 |
| 21 | 9.6 | 16.0 | 34.6 | 37.1 | 76.8 |
| end | 38.8 | 30.0 | 17.5 | 23.6 | 180.8 |
| Time | Index | Experimental Conditions | ||||
|---|---|---|---|---|---|---|
| Control | CIP_5 | Mix_2.5 | Mix_5 | Mix_10 | ||
| day 15 | H’ | 7.13 | 6.25 | 6.05 | 6.13 | 6.06 |
| E | 0.76 | 0.71 | 0.73 | 0.72 | 0.72 | |
| Chao1 | 358.95 | 281.60 | 223.17 | 259.10 | 233.50 | |
| day 21 | H’ | 6.84 | 6.47 | 6.18 | 6.17 | 6.06 |
| E | 0.75 | 0.75 | 0.73 | 0.72 | 0.72 | |
| Chao1 | 324.80 | 264.50 | 265.83 | 265.15 | 251.55 | |
| end | H’ | 6.86 | 6.24 | 6.17 | 6.23 | 5.88 |
| E | 0.76 | 0.70 | 0.70 | 0.71 | 0.66 | |
| Chao1 | 322.41 | 287.36 | 282.19 | 279.00 | 287.53 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miritana, V.M.; Patrolecco, L.; Barra Caracciolo, A.; Visca, A.; Piccinini, F.; Signorini, A.; Rosa, S.; Grenni, P.; Garbini, G.L.; Spataro, F.; et al. Effects of Ciprofloxacin Alone or in Mixture with Sulfamethoxazole on the Efficiency of Anaerobic Digestion and Its Microbial Community. Antibiotics 2022, 11, 1111. https://doi.org/10.3390/antibiotics11081111
Miritana VM, Patrolecco L, Barra Caracciolo A, Visca A, Piccinini F, Signorini A, Rosa S, Grenni P, Garbini GL, Spataro F, et al. Effects of Ciprofloxacin Alone or in Mixture with Sulfamethoxazole on the Efficiency of Anaerobic Digestion and Its Microbial Community. Antibiotics. 2022; 11(8):1111. https://doi.org/10.3390/antibiotics11081111
Chicago/Turabian StyleMiritana, Valentina Mazzurco, Luisa Patrolecco, Anna Barra Caracciolo, Andrea Visca, Flavia Piccinini, Antonella Signorini, Silvia Rosa, Paola Grenni, Gian Luigi Garbini, Francesca Spataro, and et al. 2022. "Effects of Ciprofloxacin Alone or in Mixture with Sulfamethoxazole on the Efficiency of Anaerobic Digestion and Its Microbial Community" Antibiotics 11, no. 8: 1111. https://doi.org/10.3390/antibiotics11081111
APA StyleMiritana, V. M., Patrolecco, L., Barra Caracciolo, A., Visca, A., Piccinini, F., Signorini, A., Rosa, S., Grenni, P., Garbini, G. L., Spataro, F., Rauseo, J., & Massini, G. (2022). Effects of Ciprofloxacin Alone or in Mixture with Sulfamethoxazole on the Efficiency of Anaerobic Digestion and Its Microbial Community. Antibiotics, 11(8), 1111. https://doi.org/10.3390/antibiotics11081111