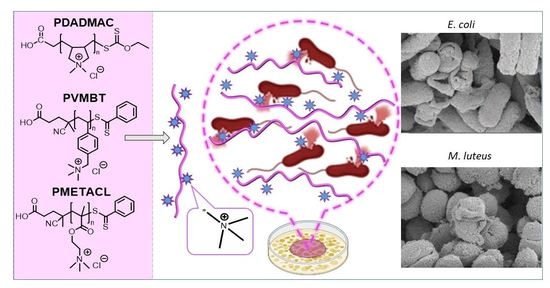
Size-Controlled Ammonium-Based Homopolymers as Broad-Spectrum Antibacterials
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of the Homopolymers
2.2. Minimum Inhibitory Concentrations of the Polymers
2.3. Investigation of the Molecular Weight and Antimicrobial Activity
2.4. Antimicrobial Mechanisms of the Polymers
2.5. Membrane Depolarization Assays
2.6. Cytotoxicity and Hemolytic Activity of Polymers
3. Materials and Methods
3.1. Methods
3.1.1. General Synthesis of Homopolymers via Reversible Addition Fragmentation Chain
Transfer Polymerization
3.1.2. P1-RAFT Polymerization of Poly(2-(dimethylamino)ethyl methacrylate)
3.1.3. P2-RAFT Polymerization of Poly([2-(methacryloyloxy)ethyl]trimethylammonium chloride)
3.1.4. P3-RAFT Polymerization of Poly[3-(methacryloylamino)propyl]trimethylammonium chloride
3.1.5. P4-RAFT Polymerization of Poly(vinylbenzyl trimethylammonium chloride)
3.1.6. P5-RAFT Polymerization of Poly(diallydimethyl ammonium chloride)
3.1.7. Synthesis of Chain Transfer Agent S-Ethoxythiocarbonyl Mercaptoacetic Acid (CTA3)
3.2. Characterization of Homopolymers
3.2.1. Nuclear Magnetic Resonance Spectroscopy
3.2.2. Molecular Weight Determination of Polymers by GPC analysis
3.3. Antimicrobial Activity of the Polymers
3.3.1. Primary Screening: Determination of Minimum Inhibitory Concentration
3.3.2. Effect of Molecular Weight on Antimicrobial Activity of Polymers
3.4. Live/Dead Assay
3.5. Bacterial Membrane Integrity Assays
3.6. Scanning Electron Microscopy
3.7. MTT Assay
3.8. Hemolysis Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Collabrators, A.R. Articles Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Wang, X.; Jing, S.; Liu, Y.; Liu, S.; Tan, Y. Diblock Copolymer Containing Bioinspired Borneol and Dopamine Moieties: Synthesis and Antibacterial Coating Applications. Polymer 2017, 116, 314–323. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Dang, F.; Ferrell, A.; Barton, H.A.; Joy, A. Peptidomimetic Polyurethanes Inhibit Bacterial Biofilm Formation and Disrupt Surface Established Biofilms. JACS 2021, 143, 9440–9449. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Yang, Y.; Yang, W.; Wang, X.; Liu, X.; Zhou, F.; Zhao, Y. One-Step Zwitterionization and Quaternization of Thick PDMAEMA Layer Grafted through Subsurface-Initiated ATRP for Robust Antibiofouling and Antibacterial Coating on PDMS. J. Colloid Interface Sci. 2022, 610, 234–245. [Google Scholar] [CrossRef] [PubMed]
- Tabriz, A.; Azeem, M.; Rehman, U.; Bilal, M.; Niazi, K. Quaternized Trimethyl Functionalized Chitosan Based Antifungal Membranes for Drinking Water Treatment Quaternized Trimethyl Functionalized Chitosan Based Antifungal Membranes for Drinking Water Treatment. Carbohydr. Polym. 2018, 207, 17–25. [Google Scholar] [CrossRef]
- Wulandari, E.; Budhisatria, R.; Soeriyadi, A.H.; Willcox, M.; Boyer, C.; Wong, E.H.H. Polymer Chemistry Combating Multidrug-Resistant Bacteria. Polym. Chem. 2021, 12, 7038–7047. [Google Scholar] [CrossRef]
- Leong, J.; Shi, D.; Pang, J.; Tan, K.; Yang, C.; Yang, S.; Wang, Y.; Ngow, Y.S.; Kng, J.; Balakrishnan, N.; et al. Potent Antiviral and Antimicrobial Polymers as Safe and Effective Disinfectants for the Prevention of Infections. Adv. Healthc. Mater. 2022, 11, 2101898. [Google Scholar] [CrossRef]
- Maria, O.; Keshari, S.; Dash, M.; Lupascu, F.; Pânzariu, A.; Tuchilus, C.; Ghetu, N.; Danciu, M.; Dubruel, P.; Pieptu, D.; et al. New Antimicrobial Chitosan Derivatives for Wound Dressing Applications. Carbohydr. Polym. 2016, 141, 28–40. [Google Scholar] [CrossRef]
- Dundas, A.A.; Sanni, O.; Dubern, J.; Dimitrakis, G.; Hook, A.L.; Irvine, D.J.; Williams, P.; Alexander, M.R. Validating a Predictive Structure—Property Relationship by Discovery of Novel Polymers Which Reduce Bacterial Biofilm Formation. Adv. Mater. 2019, 31, 1903513. [Google Scholar] [CrossRef]
- Mukherjee, S.; Barman, S.; Mukherjee, R.; Haldar, J. Amphiphilic Cationic Macromolecules Highly Effective Against Multi-Drug Resistant Gram-Positive Bacteria and Fungi With No Detectable Resistance. Front. Bioeng. Biotechnol. 2020, 8, 55. [Google Scholar] [CrossRef]
- Fu, Y.; Yang, Y.; Xiao, S.; Zhang, L.; Huang, L.; Chen, F.; Fan, P.; Zhong, M.; Tan, J.; Yang, J. Mixed Polymer Brushes with Integrated Antibacterial and Antifouling Properties. Prog. Org. Coatings 2019, 130, 75–82. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Zhang, P.; Yuan, L.; Yu, Q.; Chen, H. High Antibacterial Efficiency of PDMAEMA Modified Silicon Nanowire Arrays. Colloids Surf. B Biointerfaces 2011, 83, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Haktaniyan, M.; Bradley, M. Polymers Showing Intrinsic Antimicrobial Activity. Chem. Soc. Rev. 2022, 51, 8584–8611. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Koh, J.J.; Liu, S.; Lakshminarayanan, R.; Verma, C.S.; Beuerman, R.W. Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci. 2017, 11, 73. [Google Scholar] [CrossRef] [PubMed]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Huang, K.C.; Mukhopadhyay, R.; Wen, B.; Gitai, Z.; Wingreen, N.S. Cell Shape and Cell-Wall Organization in Gram-Negative Bacteria. Biophys. Comput. Biol. 2008, 105, 19282–19287. [Google Scholar] [CrossRef]
- Brown, S.; Santa Maria, J.P.; Walker, S. Wall Teichoic Acids of Gram-Positive Bacteria. Annu. Rev. Microbiol. 2013, 67, 313–336. [Google Scholar] [CrossRef]
- Schneewind, O.; Missiakas, D. Lipoteichoic Acids, Phosphate-Containing Polymers in the Envelope of Gram-Positive Bacteria. J. Bacteriol. 2014, 196, 1133–1142. [Google Scholar] [CrossRef]
- Khondker, A.; Rheinstädter, M.C. How Do Bacterial Membranes Resist Polymyxin Antibiotics? Commun. Biol. 2020, 3, 77. [Google Scholar] [CrossRef]
- Jiang, X.; Yang, K.; Yuan, B.; Han, M.; Zhu, Y.; Roberts, K.D.; Patil, N.A.; Li, J.; Gong, B.; Hancock, R.E.W.; et al. Molecular Dynamics Simulations Informed by Membrane Lipidomics Reveal the Structure-Interaction Relationship of Polymyxins with the Lipid A-Based Outer Membrane of Acinetobacter Baumannii. J. Antimicrob. Chemother. 2020, 75, 3534–3543. [Google Scholar] [CrossRef]
- Yang, X.; Huang, E.; Yousef, A.E. Brevibacillin, a Cationic Lipopeptide That Binds to Lipoteichoic Acid and Subsequently Disrupts Cytoplasmic Membrane of Staphylococcus Aureus. Microbiol. Res. 2017, 195, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Heesterbeek, D.A.C.; Martin, N.I.; Velthuizen, A.; Duijst, M.; Ruyken, M.; Wubbolts, R.; Rooijakkers, S.H.M.; Bardoel, B.W. Complement-Dependent Outer Membrane Perturbation Sensitizes Gram-Negative Bacteria to Gram-Positive Specific Antibiotics. Sci. Rep. 2019, 9, 3074. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.R.; Fonseca, A.C.; Mendonça, P.V.; Branco, R.; Serra, A.C.; Morais, P.V.; Coelho, J.F. Recent Developments in Antimicrobial Polymers: A Review. Materials 2016, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Dias, F.G.G.; de Freitas Pereira, L.; Parreira, R.L.T.; Veneziani, R.C.S.; Bianchi, T.C.; de Paula Fontes, V.F.N.; de Carlos Galvani, M.; Cerce, D.D.P.; Martins, C.H.G.; Rinaldi-Neto, F.; et al. Evaluation of the Antiseptic and Wound Healing Potential of Polyhexamethylene Guanidine Hydrochloride as Well as Its Toxic Effects. Eur. J. Pharm. Sci. 2021, 160, 105739. [Google Scholar] [CrossRef]
- Kumar, S.; Pillai, R.; Reghu, S.; Vikhe, Y.; Zheng, H.; Koh, C.H.; Chan-park, M.B. Novel Antimicrobial Coating on Silicone Contact Lens Using Glycidyl Methacrylate and Polyethyleneimine Based Polymers. Macromol. Rapid Commun. 2020, 41, 20000175. [Google Scholar] [CrossRef]
- Gultekinoglu, M.; Karahan, S.; Kart, D.; Sagiroglu, M.; Erta, N.; Ozen, A.H.; Ulubayram, K. Polyethyleneimine Brushes Effectively Inhibit Encrustation on Polyurethane Ureteral Stents Both in Dynamic Bioreactor and in Vivo. Mater. Sci. Eng. C 2017, 71, 1166–1174. [Google Scholar] [CrossRef]
- Peng, J.; Liu, P.; Peng, W.; Sun, J.; Dong, X.; Ma, Z. Poly (Hexamethylene Biguanide) (PHMB) as High-Efficiency Antibacterial Coating for Titanium Substrates. J. Hazard. Mater. 2021, 411, 125110. [Google Scholar] [CrossRef]
- Rahman, M.A.; Jui, M.S.; Bam, M.; Cha, Y.; Luat, E.; Alabresm, A.; Nagarkatti, M.; Decho, A.W.; Tang, C. Facial Amphiphilicity-Induced Polymer Nanostructures for Antimicrobial Applications. ACS Appl. Mater. Interfaces 2020, 12, 21221–21230. [Google Scholar] [CrossRef]
- Venkateswaran, S.; Wu, M.; Gwynne, P.J.; Hardman, A.; Lilienkampf, A.; Pernagallo, S.; Blakely, G.; Swann, D.G.; Gallagher, M.P.; Bradley, M. Bacteria Repelling Poly(Methylmethacrylate-Co-Dimethylacrylamide) Coatings for Biomedical Devices Devices. J. Mater. Chem. B 2014, 2, 6723–6729. [Google Scholar] [CrossRef]
- Dizman, B.; Elasri, M.O.; Mathias, L.J. Synthesis and Characterization of Antibacterial and Temperature Responsive Methacrylamide Polymers. Macromolecules 2006, 39, 5738–5746. [Google Scholar] [CrossRef]
- Hoque, J.; Akkapeddi, P.; Yadav, V.; Manjunath, G.B.; Uppu, D.S.S.M.; Konai, M.M.; Yarlagadda, V.; Sanyal, K.; Haldar, J. Broad Spectrum Antibacterial and Antifungal Polymeric Paint Materials: Synthesis, Structure—Activity Relationship, and Membrane-Active Mode of Action. Appl. Mater. 2015, 7, 1804–1815. [Google Scholar] [CrossRef]
- Wee, V.; Ng, L.; Pang, J.; Tan, K.; Leong, J.; Voo, Z.X.; Hedrick, J.L. Antimicrobial Polycarbonates: Investigating the Impact of Nitrogen- Containing Heterocycles as Quaternizing Agents. Macromolecules 2014, 47, 1285–1291. [Google Scholar]
- Oh, J.; Kim, S.; Oh, M.; Khan, A. Antibacterial Properties of Main-Chain Cationic Polymers Prepared through Amine–Epoxy ‘Click’ Polymerization. RSC Adv. 2020, 10, 26752–26755. [Google Scholar] [CrossRef] [PubMed]
- Yandi, W.; Mieszkin, S.; Callow, M.E.; Callow, J.A.; Finlay, J.A.; Liedberg, B.; Ederth, T. Antialgal Activity of Poly(2-(Dimethylamino)Ethyl Methacrylate) (PDMAEMA) Brushes against the Marine Alga Ulva. Biofouling 2017, 33, 169–183. [Google Scholar] [CrossRef]
- Ji, W.; Koepsel, R.R.; Murata, H.; Zadan, S.; Campbell, A.S.; Russell, A.J. Bactericidal Specificity and Resistance Profile of Poly(Quaternary Ammonium) Polymers and Protein-Poly(Quaternary Ammonium) Conjugates. Biomacromolecules 2017, 18, 2583–2593. [Google Scholar] [CrossRef]
- Phillips, D.J.; Harrison, J.; Richards, S.J.; Mitchell, D.E.; Tichauer, E.; Hubbard, A.T.M.; Guy, C.; Hands-Portman, I.; Fullam, E.; Gibson, M.I. Evaluation of the Antimicrobial Activity of Cationic Polymers against Mycobacteria: Toward Antitubercular Macromolecules. Biomacromolecules 2017, 18, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Cai, Z.; Huang, Z.; Tang, X.; Zhang, X. Antimicrobial Cationic Polymers: From Structural Design to Functional Control. Polym. J. 2018, 50, 33–44. [Google Scholar] [CrossRef]
- De Jesús-Téllez, M.A.; De la Rosa-García, S.; Medrano-Galindo, I.; Rosales-Peñafiel, I.; Gómez-Cornelio, S.; Guerrero-Sanchez, C.; Schubert, U.S.; Quintana-Owen, P. Antifungal Properties of Poly[2-(Dimethylamino)Ethyl Methacrylate] (PDMAEMA) and Quaternized Derivatives. React. Funct. Polym. 2021, 163, 104887. [Google Scholar] [CrossRef]
- Keum, H.; Kim, D.; Whang, C.-H.; Kang, A.; Lee, S.; Na, W.; Jon, S. Impeding the Medical Protective Clothing Contamination by a Spray Coating of Trifunctional Polymers. ACS Omega 2022, 7, 10526–10538. [Google Scholar] [CrossRef]
- Qiu, H.; Si, Z.; Luo, Y.; Feng, P.; Wu, X.; Hou, W.; Zhu, Y.; Chan-Park, M.B.; Xu, L.; Huang, D. The Mechanisms and the Applications of Antibacterial Polymers in Surface Modification on Medical Devices. Front. Bioeng. Biotechnol. 2020, 8, 910. [Google Scholar] [CrossRef]
- Kwaśniewska, D.; Chen, Y.L.; Wieczorek, D. Biological Activity of Quaternary Ammonium Salts and Their Derivatives. Pathogens 2020, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Hirayama, H.; Yamaguchi, H.; Tazuke, S. Polycationic Biocides with Pendant Active Groups: Molecular Weight Dependence of Antibacterial Activity. Antimicrob. Agents Chemother. 1986, 30, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Hj, M.Á. The Effect of Molecular Weight on the Antibacterial Activity of N,N,N-Trimethyl Chitosan (TMC). Int. J. Mol. Sci. 2019, 20, 1743. [Google Scholar] [CrossRef]
- Guo, J.; Qin, J.; Ren, Y.; Wang, B.; Cui, H.; Ding, Y.; Mao, H.; Yan, F. Antibacterial Activity of Cationic Polymers: Side-Chain or Main-Chain Type? Polym. Chem. 2018, 9, 4611–4616. [Google Scholar] [CrossRef]
- Santos, M.R.E.; Mendonça, P.V.; Almeida, M.C.; Branco, R.; Serra, A.C.; Morais, P.V.; Coelho, J.F.J. Increasing the Antimicrobial Activity of Amphiphilic Cationic Copolymers by the Facile Synthesis of High Molecular Weight Stars by Supplemental Activator and Reducing Agent Atom Transfer Radical Polymerization. Biomacromolecules 2019, 20, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Pham, P.; Oliver, S.; Wong, E.H.H.; Boyer, C. Effect of Hydrophilic Groups on the Bioactivity of Antimicrobial Polymers. Polym. Chem. 2021, 12, 5689–5703. [Google Scholar] [CrossRef]
- Phuong, P.T.; Oliver, S.; He, J.; Wong, E.H.H.; Mathers, R.T.; Boyer, C. Effect of Hydrophobic Groups on Antimicrobial and Hemolytic Activity: Developing a Predictive Tool for Ternary Antimicrobial Polymers. Biomacromolecules 2020, 21, 5241–5255. [Google Scholar] [CrossRef]
- Copolymer, P.; Xue, Y.; Xiao, H. Antibacterial/Antiviral Property and Mechanism of Dual-Functional Quaternized Pyridinium-Type Copolymer. Polymers 2015, 7, 2290–2303. [Google Scholar] [CrossRef]
- Roka, N.; Kokkorogianni, O.; Kontoes-Georgoudakis, P.; Choinopoulos, I.; Pitsikalis, M. Recent Advances in the Synthesis of Complex Macromolecular Architectures Based on Poly(N-Vinyl Pyrrolidone) and the RAFT Polymerization Technique. Polymers 2022, 14, 701. [Google Scholar] [CrossRef]
- Nothling, M.D.; Fu, Q.; Reyhani, A.; Allison-Logan, S.; Jung, K.; Zhu, J.; Kamigaito, M.; Boyer, C.; Qiao, G.G. Progress and Perspectives Beyond Traditional RAFT Polymerization. Adv. Sci. 2020, 7, 2001656. [Google Scholar] [CrossRef]
- Paslay, L.C.; Abel, B.A.; Brown, T.D.; Koul, V.; Choudhary, V.; McCormick, C.L.; Morgan, S.E. Antimicrobial Poly(Methacrylamide) Derivatives Prepared via Aqueous RAFT Polymerization Exhibit Biocidal Efficiency Dependent upon Cation Structure. Biomacromolecules 2012, 13, 2472–2482. [Google Scholar] [CrossRef] [PubMed]
- Sathyan, A.; Kurtz, I.; Rathore, P.; Emrick, T.; Schiffman, J.D. Using Catechol and Zwitterion-Functionalized Copolymers to Prevent Dental Bacterial Adhesion. ACS Appl. Bio Mater. 2023, 6, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Song, F.; Zhang, L.; Chen, R.; Liu, Q.; Liu, J.; Yu, J.; Liu, P.; Duan, J.; Wang, J. Bioinspired Durable Antibacterial and Antifouling Coatings Based on Borneol Fluorinated Polymers: Demonstrating Direct Evidence of Antiadhesion. ACS Appl. Mater. Interfaces 2021, 13, 33417–33426. [Google Scholar] [CrossRef]
- Keely, S.; Rawlinson, L.A.B.; Haddleton, D.M.; Brayden, D.J. A Tertiary Amino-Containing Polymethacrylate Polymer Protects Mucus-Covered Intestinal Epithelial Monolayers against Pathogenic Challenge. Pharm. Res. 2008, 25, 1193–1201. [Google Scholar] [CrossRef]
- Rawlinson, L.A.B.; O’Gara, J.P.; Jones, D.S.; Brayden, D.J. Resistance of Staphylococcus Aureus to the Cationic Antimicrobial Agent Poly(2-(Dimethylamino Ethyl)Methacrylate) (PDMAEMA) Is Influenced by Cell-Surface Charge and Hydrophobicity. J. Med. Microbiol. 2011, 60, 968–976. [Google Scholar] [CrossRef] [PubMed]
- Vitro, I.; Skóra, M. Studies on Antifungal Properties of Methacrylamido Propyl Trimethyl Ammonium Chloride Polycations and Their Toxicity. Microbiol. Spectr. 2023, 11, e0084423. [Google Scholar] [CrossRef]
- Grace, J.L.; Huang, J.X.; Cheah, S.E.; Truong, N.P.; Cooper, M.A.; Li, J.; Davis, T.P.; Quinn, J.F.; Velkov, T.; Whittaker, M.R. Antibacterial Low Molecular Weight Cationic Polymers: Dissecting the Contribution of Hydrophobicity, Chain Length and Charge to Activity. RSC Adv. 2016, 6, 15469–15477. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Wu, J.H.; Jia, H.Q.; Hao, L.M.; Wang, R.Z.; Qi, J.C. Facile Preparation and Antibacterial Properties of Cationic Polymers Derived from 2-(Dimethylamino)Ethyl Methacrylate. RSC Adv. 2013, 3, 20758–20764. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Cai, J.; Yue, Y.; Rui, D.; Zhang, Y.; Liu, S.; Wu, C. Effect of Chain Length on Cytotoxicity and Endocytosis of Cationic Polymers. Macromol. Res. 2011, 44, 2050–2057. [Google Scholar] [CrossRef]
- Correia, J.S.; Mirón-barroso, S.; Hutchings, C.; Ottaviani, S.; Castellano, L. How Does the Polymer Architecture and Position of Cationic Charges Affect Cell Viability? Polym. Chem. 2022, 14, 303–317. [Google Scholar] [CrossRef] [PubMed]
- Online, V.A. A Polyion Complex Micelle with Heparin for Growth Factorfactor Delivery and Uptake into Cells. J. Mater. Chem. B 2013, 1, 1635–1643. [Google Scholar] [CrossRef]
- Monnery, B.D.; Wright, M.; Cavill, R.; Hoogenboom, R.; Shaunak, S.; Steinke, J.H.G.; Thanou, M. Cytotoxicity of Polycations: Relationship of Molecular Weight and the Hydrolytic Theory of the Mechanism of Toxicity. Int. J. Pharm. 2017, 521, 249–258. [Google Scholar] [CrossRef]
- Locock, K.E.S.; Michl, T.D.; Valentin, J.D.P.; Vasilev, K.; Hayball, J.D.; Qu, Y.; Traven, A.; Griesser, H.J.; Meagher, L.; Haeussler, M. Guanylated Polymethacrylates: A Class of Potent Antimicrobial Polymers with Low Hemolytic Activity. Biomacromolecules 2013, 14, 4021–4031. [Google Scholar] [CrossRef] [PubMed]
- Layman, J.M.; Ramirez, S.M.; Green, M.D.; Long, T.E. Influence of Polycation Molecular Weight on Poly(2-Dimethylaminoethyl Methacrylate)-Mediated DNA Delivery in Vitro. Biomacromolecules 2009, 10, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Samsonova, O.; Pfeiffer, C.; Hellmund, M.; Merkel, O.M.; Kissel, T. Low Molecular Weight PDMAEMA-Block-PHEMA Block-Copolymers Synthesized via RAFT-Polymerization: Potential Non-Viral Gene Delivery Agents. Polymers 2011, 3, 693–718. [Google Scholar] [CrossRef]
- Foster, L.L.; Yusa, S.I.; Kuroda, K. Solution-Mediated Modulation of Pseudomonas Aeruginosa Biofilm Formation by a Cationic Synthetic Polymer. Antibiotics 2019, 8, 61. [Google Scholar] [CrossRef]
- Perrier, S. 50th Anniversary Perspective: RAFT Polymerization-A User Guide. Macromolecules 2017, 50, 7433–7447. [Google Scholar] [CrossRef]
- Cao, J.; Siefker, D.; Chan, B.A.; Yu, T.; Lu, L.; Saputra, M.A.; Fronczek, F.R.; Xie, W.; Zhang, D. Interfacial Ring-Opening Polymerization of Amino-Acid-Derived N-Thiocarboxyanhydrides toward Well-Defined Polypeptides. ACS Macro Lett. 2017, 6, 836–840. [Google Scholar] [CrossRef]
- Druvari, D.; Koromilas, N.D.; Bekiari, V.; Bokias, G.; Kallitsis, J.K. Polymeric Antimicrobial Coatings Based on Quaternary Ammonium Compounds. Coatings 2018, 8, 8. [Google Scholar] [CrossRef]
- Kenawy, E.R.; Worley, S.D.; Broughton, R. The Chemistry and Applications of Antimicrobial Polymers: A State-of-the-Art Review. Biomacromolecules 2007, 8, 1359–1384. [Google Scholar] [CrossRef] [PubMed]
- Demarteau, J.; De Añastro, F.; Shaplov, A.S. Polymer Chemistry. Polym. Chem. 2020, 11, 1481–1488. [Google Scholar] [CrossRef]
- Teh, C.H.; Nazni, W.A.; Nurulhusna, A.H.; Norazah, A.; Lee, H.L. Determination of Antibacterial Activity and Minimum Inhibitory Concentration of Larval Extract of Fly via Resazurin-Based Turbidometric Assay. BMC Microbiol. 2017, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Barman, R.; Das, B.; Ghosh, S. Highly Efficient Biofilm Eradication by Antibacterial Two-Dimensional Supramolecular Polymers. Chem. Mater. 2021, 33, 8656–8665. [Google Scholar] [CrossRef]
- Wu, C.L.; Peng, K.L.; Yip, B.S.; Chih, Y.H.; Cheng, J.W. Boosting Synergistic Effects of Short Antimicrobial Peptides with Conventional Antibiotics against Resistant Bacteria. Front. Microbiol. 2021, 12, 747760. [Google Scholar] [CrossRef]
- Singhsa, P.; Diaz-Dussan, D.; Manuspiya, H.; Narain, R. Well-Defined Cationic N-[3-(Dimethylamino)Propyl]Methacrylamide Hydrochloride-Based (Co)Polymers for SiRNA Delivery. Biomacromolecules 2018, 19, 209–221. [Google Scholar] [CrossRef]
- Azuma, R.; Nakamichi, S.; Kimura, J.; Yano, H.; Kawasaki, H.; Suzuki, T.; Kondo, R.; Kanda, Y.; Shimizu, K.I.; Kato, K.; et al. Solution Synthesis of N,N-Dimethylformamide-Stabilized Iron-Oxide Nanoparticles as an Efficient and Recyclable Catalyst for Alkene Hydrosilylation. ChemCatChem 2018, 10, 2378–2382. [Google Scholar] [CrossRef]







| Code | Polymer | CTA | [M]/[I] | [CTA]/[I] | Mn Calc. | Mn-GPC | Mw-GPC | PDI |
|---|---|---|---|---|---|---|---|---|
| P1 | PDMAEMA | 1 | 167 | 6 | 23.0 kDa | 22 kDa | 28 kDa | 1.29 |
| P2 | PMETACL | 2 | 100 | 5 | 17.1 kDa | 17 kDa | 20 kDa | 1.19 |
| P3 | PMET3 | 2 | 215 | 5 | 23.1 kDa | 14 kDa | 16 kDa | 1.18 |
| P4 | PVMBT | 2 | 190 | 7 | 20.8 kDa | 14 kDa | 16 kDa | 1.11 |
| P5 | PDADMAC | 3 | 175 | 3.3 | 17.5 kDa | 18 kDa | 24 kDa | 1.36 |
| Polymer | Code | MICs for Different Target Microorganisms | |||||||
|---|---|---|---|---|---|---|---|---|---|
| B. subtilis | E. coli | M. luteus | S. typhimurium | ||||||
| µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/ mL | µM | ||
| PDMAEMA | P1 | 32 | 1.6 | 32 | 1.6 | 32 | 1.6 | 32 | 1.6 |
| PMETACL | P2 | 64 | 1.9 | 64 | 1.9 | 64 | 1.9 | 64 | 1.9 |
| PMET3 | P3 | 32 | 1.4 | 32 | 1.5 | 16 | 0.7 | 64 | 2.8 |
| PVBMT | P4 | 32 | 0.9 | 64 | 1.7 | 64 | 1.7 | 64 | 1.7 |
| PDADMAC | P5 | 16 | 0.9 | 16 | 0.9 | 16 | 0.9 | 32 | 1.8 |
| Standard Antibiotics | Clindamycin | Gentamicin | Gentamicin | Chloramphenicol | |||||
| 4 | 9.4 | 0.5 | 1.1 | 0.5 | 1.1 | 1 | 3.1 | ||
| Polymer | Mn Calc. | Mn-GPC | Mw-GPC | PDI |
|---|---|---|---|---|
| PMETACI-10 | 10.4 kDa | 11 kDa | 14 kDa | 1.24 |
| PMETACI-20 | 17.1 kDa | 17 kDa | 20 kDa | 1.19 |
| PMETACI-40 | 39.7 kDa | 39 kDa | 40 kDa | 1.03 |
| PDADMAC-10 | 11.9 kDa | 11 kDa | 15 kDa | 1.36 |
| PDADMAC-20 | 17.5 kDa | 18 kDa | 24 kDa | 1.36 |
| PDADMAC-40 | 36.8 kDa | 35 kDa | 48 kDa | 1.37 |
| PVMBT-10 | 13.7 kDa | 6 kDa | 7 kDa | 1.20 |
| PVMBT-20 | 20.8 kDa | 14 kDa | 16 kDa | 1.11 |
| PVMBT-40 | 37.4 kDa | 23 kDa | 27 kDa | 1.18 |
| Polymer Mn | Polymer Name | MICs for Different Target Microorganisms | |||||||
|---|---|---|---|---|---|---|---|---|---|
| B. subtilis | E. coli | M. luteus | S. typhimurium | ||||||
| µg/mL | µM | µg/mL | µM | µg/mL | µM | µg/ mL | µM | ||
| 10 kDa | PDADMAC-10 | 16 | 1.4 | 32 | 2.7 | 32 | 2.7 | 64 | 5.4 |
| 17 kDa | PDADMAC-20 | 16 | 0.9 | 16 | 0.9 | 16 | 0.9 | 32 | 1.8 |
| 40 kDa | PDADMAC-40 | 16 | 0.4 | 16 | 0.4 | 16 | 0.4 | 32 | 0.9 |
| 12 kDa | PMETACL-10 | 64 | 6.2 | 64 | 6.2 | 64 | 6.2 | 64 | 6.2 |
| 18 kDa | PMETACL-20 | 64 | 3.7 | 64 | 3.7 | 64 | 3.7 | 64 | 3.7 |
| 37 kDa | PMETACL-40 | 32 | 0.9 | 32 | 0.9 | 32 | 0.9 | 64 | 1.7 |
| 14 kDa | PVMBT-10 | 64 | 4.7 | 64 | 4.7 | 64 | 4.7 | 128 | 9.3 |
| 21 kDa | PVMBT-20 | 32 | 1.5 | 32 | 1.5 | 64 | 3.1 | 64 | 3.1 |
| 37 kDa | PVMBT-40 | 32 | 0.9 | 32 | 10.9 | 32 | 0.9 | 64 | 1.7 |
| Polymer | MIC (µg/mL) | HC50 mg/mL | Selectivity HC50/MIC | |||
|---|---|---|---|---|---|---|
| E. coli | B. subtilis | M. luteus | S.typhimurium | |||
| PDADMAC-20 | 16 | 16 | 16 | 32 | >26 | >800 |
| PVMBT-40 | 32 | 32 | 32 | 64 | >51 | >800 |
| PMETACL-40 | 32 | 32 | 32 | 64 | >51 | >800 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haktaniyan, M.; Sharma, R.; Bradley, M. Size-Controlled Ammonium-Based Homopolymers as Broad-Spectrum Antibacterials. Antibiotics 2023, 12, 1320. https://doi.org/10.3390/antibiotics12081320
Haktaniyan M, Sharma R, Bradley M. Size-Controlled Ammonium-Based Homopolymers as Broad-Spectrum Antibacterials. Antibiotics. 2023; 12(8):1320. https://doi.org/10.3390/antibiotics12081320
Chicago/Turabian StyleHaktaniyan, Meltem, Richa Sharma, and Mark Bradley. 2023. "Size-Controlled Ammonium-Based Homopolymers as Broad-Spectrum Antibacterials" Antibiotics 12, no. 8: 1320. https://doi.org/10.3390/antibiotics12081320
APA StyleHaktaniyan, M., Sharma, R., & Bradley, M. (2023). Size-Controlled Ammonium-Based Homopolymers as Broad-Spectrum Antibacterials. Antibiotics, 12(8), 1320. https://doi.org/10.3390/antibiotics12081320