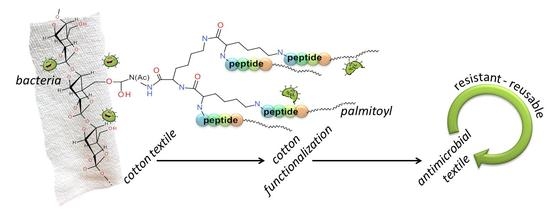
Covalent Graft of Lipopeptides and Peptide Dendrimers to Cellulose Fibers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Cotton Functionalization
2.3. Peptide Synthesis on the Functionalized Cotton
2.4. Characterization of the Peptide-Cotton Samples by X-Ray Photoelectron Spectroscopy (XPS)
2.5. Evaluation of the Whiteness of the Functionalized Cotton Samples
2.6. Antimicrobial Activity
3. Results and Discussion
3.1. Peptide Design
3.2. Synthesis of the Peptide-Cotton Samples
3.3. Peptide Loading Determination and FT-IR Absorption Analysis
3.4. XPS Analysis
3.5. Color Change and Antimicrobial Activity
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gao, Y.; Cranston, R. Recent advances in antimicrobial treatments of textiles. Text. Res. J. 2008, 78, 60–72. [Google Scholar] [CrossRef]
- Morais, D.S.; Guedes, R.M.; Lopes, M.A. Antimicrobial approaches for textiles: From research to market. Materials 2016, 9, 498. [Google Scholar] [CrossRef] [PubMed]
- Tawiah, B.; Badoe, W.; Fu, S.H. Advances in the development of antimicrobial agents for textiles: The quest for natural products. Review. Fibres Text. East Eur. 2016, 24, 136–149. [Google Scholar] [CrossRef]
- Wang, W.Y.; Hui, P.C.L.; Kan, C.W. Functionalized textile based therapy for the treatment of atopic dermatitis. Coatings 2017, 7, 82. [Google Scholar] [CrossRef]
- Zhou, C.E.; Kan, C.W.; Matinlinna, J.P.; Tsoi, J.K.H. Regenerable antibacterial cotton fabric by plasma treatment with dimethylhydantoin: Antibacterial activity against S. aureus. Coatings 2017, 7, 11. [Google Scholar] [CrossRef]
- Cardoso, V.; Rittmeyer, T.; Correa, R.J.; Brêda, G.C.; Almeida, R.V.; Simões, G.; de França, B.M.; de Azevedo, P.N.; Forero, J.S.B. Photoactive cotton fabric: Synthesis, characterization and antibacterial evaluation of anthraquinone-based dyes linked to cellulose. Dye. Pigment. 2019, 161, 16–23. [Google Scholar] [CrossRef]
- Marković, D.; Deeks, C.; Nunney, T.; Radovanović, Ž.; Radoičić, M.; Šaponjić, Z.; Radetić, M. Antibacterial activity of Cu-based nanoparticles synthesized on the cotton fabrics modified with polycarboxylic acids. Carbohyd. Polym. 2018, 200, 173–182. [Google Scholar] [CrossRef]
- Yaghoubidoust, F.; Salimi, E. A simple method for the preparation of antibacterial cotton fabrics by coating graphene oxide nanosheets. Fiber Polym. 2019, 20, 1155–1160. [Google Scholar] [CrossRef]
- Stular, D.; Sobak, M.; Mihelcic, M.; Sest, E.; Ilic, I.G.; Jerman, I.; Simoncic, B.; Tomsic, B. Proactive release of antimicrobial essential oil from a “smart” cotton fabric. Coatings 2019, 9, 242. [Google Scholar] [CrossRef]
- Gugliuzza, A.; Drioli, E. A review on membrane engineering for innovation in wearable fabrics and protective textiles. J. Membr. Sci. 2013, 446, 350–375. [Google Scholar] [CrossRef]
- Kenawy, E.-R.; Worley, S.D.; Broughton, R. The chemistry and applications of antimicrobial polymers: A state-of-the-art review. Biomacromolecules 2007, 8, 1359–1384. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, D.R.; Gorup, L.F.; Takamiya, A.S.; Ruvollo-Filho, A.C.; de Camargo, E.R.; Barbosa, D.B. The growing importance of materials that prevent microbial adhesion: Antimicrobial effect of medical devices containing silver. Int. J. Antimicrob. Agents 2009, 34, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Stobie, N.; Duffy, B.; Hinder, S.J.; McHale, P.; McCormack, D.E. Silver doped perfluoropolyether-urethane coatings: Antibacterial activity and surface analysis. Colloid Surf. B 2009, 72, 62–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassano, R.; Trombino, S.; Ferrarelli, T.; Barone, E.; Arena, V.; Mancuso, C.; Picci, N. Synthesis, characterization, and anti-inflammatory activity of diclofenac-bound cotton fibers. Biomacromolecules 2010, 11, 1716–1720. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, G.; Chakraborty, J.N. Antimicrobial performance of cotton finished with triclosan, silver and chitosan. Fash. Text. 2015, 2, 13. [Google Scholar] [CrossRef]
- El-Nahhal, I.M.; Zourab, S.M.; Kodeh, F.S.; Selmane, M.; Genois, I.; Babonneau, F. Nanostructured copper oxide-cotton fibers: Synthesis, characterization, and applications. Int. Nano Lett. 2012, 2, 14. [Google Scholar] [CrossRef]
- El-Nahhal, I.M.; Zourab, S.M.; Kodeh, F.S.; Elmanama, A.A.; Selmane, M.; Genois, I.; Babonneau, F. Nano-structured zinc oxide-cotton fibers: Synthesis, characterization and applications. J. Mater. Sci. Mater. Electron. 2013, 24, 3970–3975. [Google Scholar] [CrossRef]
- Ayyoob, M.; Khurshid, M.F.; Asad, M.; Shah, S.N.H. Assessment of eco-friendly natural antimicrobial textile finish extracted from aloe vera and neem plants. Fibres Text. East Eur. 2015, 23, 120–123. [Google Scholar] [CrossRef]
- Shahid-ul-Islam; Mohammad, F. Natural colorants in the presence of anchors so-called mordants as promising coloring and antimicrobial agents for textile materials. Acs Sustain. Chem. Eng. 2015, 3, 2361–2375. [Google Scholar] [CrossRef]
- Gomes, A.P.; Mano, J.F.; Queiroz, J.A.; Gouveia, I.C. Layer-by-layer deposition of antimicrobial polymers on cellulosic fibers: A new strategy to develop bioactive textiles. Polym. Adv. Technol. 2013, 24, 1005–1010. [Google Scholar] [CrossRef]
- Gomes, A.P.; Mano, J.F.; Queiroz, J.A.; Gouveia, I.C. Incorporation of antimicrobial peptides on functionalized cotton gauzes for medical applications. Carbohydr. Polym. 2015, 127, 451–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.-M.; Chen, X.; Yuan, B.-F.; Feng, Y.-Q. Graft modification of cotton with phosphate group and its application to the enrichment of phosphopeptides. J. Chromatogr. A 2017, 1484, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Hsu, B.B.; Klibanov, A.M. Light-activated covalent coating of cotton with bactericidal hydrophobic polycations. Biomacromolecules 2011, 12, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Ringot, C.; Sol, V.; Barrière, M.; Saad, N.; Bressollier, P.; Granet, R.; Couleaud, P.; Frochot, C.; Krausz, P. Triazinyl porphyrin-based photoactive cotton fabrics: Preparation, characterization, and antibacterial activity. Biomacromolecules 2011, 12, 1716–1723. [Google Scholar] [CrossRef] [PubMed]
- Muresan, E.I.; Balan, G.; Popescu, V. Durable hydrophobic treatment of cotton fabrics with glycidyl stearate. Ind. Eng. Chem. Res. 2013, 52, 6270–6276. [Google Scholar] [CrossRef]
- Edwards, J.V.; Fontenot, K.R.; Haldane, D.; Prevost, N.T.; Condon, B.D.; Grimm, C. Human neutrophil elastase peptide sensors conjugated to cellulosic and nanocellulosic materials: Part I, synthesis and characterization of fluorescent analogs. Cellulose 2016, 23, 1283–1295. [Google Scholar] [CrossRef]
- Edwards, J.V.; Mao, N.; Russell, S.; Carus, E.; Condon, B.; Hinchliffe, D.; Gary, L.; Graves, E.; Bopp, A.; Wang, Y. Fluid handling and fabric handle profiles of hydroentangled greige cotton and spunbond polypropylene nonwoven topsheets. Proc. Inst. Mech. Eng. Part L J. Mater. Des. Appl. 2016, 230, 847–859. [Google Scholar] [CrossRef]
- Muresan, E.I.; Piroi, C.; Creanga, D.; Stelea, L.; Oprica, L.; Sandu, I. Glycidyl esters used for multifunctional finishing of textile materials. Rev. Chim. Buchar. 2016, 5, 871–875. [Google Scholar]
- Edwards, J.V.; Fontenot, K.R.; Prevost, N.T.; Pircher, N.; Liebner, F.; Condon, B.D. Preparation, characterization and activity of a peptide-cellulosic aerogel protease sensor from cotton. Sensors 2016, 16, 1789. [Google Scholar] [CrossRef] [PubMed]
- Orlandin, A.; Formaggio, F.; Toffoletti, A.; Peggion, C. Cotton functionalized with peptides: Characterization and synthetic methods. J. Pept. Sci. 2014, 20, 547–553. [Google Scholar] [CrossRef]
- Dzuba, S.A.; Uvarov, M.N.; Utkin, D.E.; Formaggio, F.; Bedon, A.; Orlandin, A.; Peggion, C. The power of EPR techniques in investigating functionalization and penetration into fibers of cotton-bound antimicrobial peptides. Appl. Magn. Reson. 2017, 48, 943–953. [Google Scholar] [CrossRef]
- Nakamura, M.; Iwasaki, T.; Tokino, S.; Asaoka, A.; Yamakawa, M.; Ishibashi, J. Development of a bioactive fiber with immobilized synthetic peptides designed from the active site of a beetle defensin. Biomacromolecules 2011, 12, 1540–1545. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Matsuzaki, K. Control of cell selectivity of antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 1687–1692. [Google Scholar] [CrossRef] [Green Version]
- Haney, E.F.; Hunter, H.N.; Matsuzaki, K.; Vogel, H.J. Solution NMR studies of amphibian antimicrobial peptides: Linking structure to function? Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 1639–1655. [Google Scholar] [CrossRef] [Green Version]
- Koehbach, J.; Craik, D.J. The vast structural diversity of antimicrobial peptides. Trends Pharmacol. Sci. 2019, 7, 517–528. [Google Scholar] [CrossRef]
- Gabernet, G.; Müller, A.T.; Hiss, J.A.; Schneider, G. Membranolytic anticancer peptides. Med. Chem. Commun. 2016, 7, 2232–2245. [Google Scholar] [CrossRef]
- Dalzini, A.; Bergamini, C.; Biondi, B.; De Zotti, M.; Panighel, G.; Fato, R.; Peggion, C.; Bortolus, M.; Maniero, A.L. The rational search for selective anticancer derivatives of the peptide trichogin GA IV: A multi-technique biophysical approach. Sci. Rep. 2016, 6, 24000. [Google Scholar] [CrossRef]
- De Zotti, M.; Biondi, B.; Peggion, C.; Formaggio, F.; Park, Y.; Hahm, K.-S.; Toniolo, C. Trichogin GA IV: A versatile template for the synthesis of novel peptaibiotics. Org. Biomol. Chem. 2012, 10, 1285–1299. [Google Scholar] [CrossRef]
- Formaggio, F.; Peggion, C.; Crisma, M.; Toniolo, C. Short-chain analogues of the lipopeptaibol antibiotic trichogin GA IV: Conformational analysis and membrane modifying properties. J. Chem. Soc. Perkin Trans. 2001, 2, 1372–1377. [Google Scholar] [CrossRef]
- Hilpert, K.; Volkmer-Engert, R.; Walter, T.; Hancock, R.E. High-throughput generation of small antibacterial peptides with improved activity. Nat. Biotechnol. 2005, 23, 1008–1012. [Google Scholar] [CrossRef]
- Oancea, S.; Hilma, G.; Peggion, C.; Formaggio, F.; Toniolo, C. Main-chain length control of conformation, membrane activity, and antibiotic properties of lipopeptaibol sequential analogues. Chem. Biodivers. 2008, 5, 681–692. [Google Scholar] [CrossRef]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic, α-helical antimicrobial peptides. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef]
- Makovitzki, A.; Avrahami, D.; Shai, Y. Ultrashort antibacterial and antifungal lipopeptides. Proc. Natl. Acad. Sci. USA 2006, 103, 15997–16002. [Google Scholar] [CrossRef] [Green Version]
- Makovitzki, A.; Baram, J.; Shai, Y. Antimicrobial lipopolypeptides composed of palmitoyl di- and tricationic peptides: In vitro and in vivo activities, self-assembly to nanostructures, and a plausible mode of action. Biochemistry 2008, 47, 10630–10636. [Google Scholar] [CrossRef]
- Young, A.W.; Liu, Z.; Zhou, C.; Totsingan, F.; Jiwrajka, N.; Shi, Z.; Kallenbach, N.R. Structure and antimicrobial properties of multivalent short peptides. Med. Chem. Commun. 2011, 2, 308–314. [Google Scholar] [CrossRef]
- Findlay, B.; Zhanel, G.G.; Schweizer, F. Investigating the antimicrobial peptide ‘window of activity’ using cationic lipopeptides with hydrocarbon and fluorinated tails. Int. J. Antimicrob. Agents 2012, 40, 36–42. [Google Scholar] [CrossRef]
- Opitakorn, A.; Rauytanapanit, M.; Waditee-Sirisattha, R.; Praneenararat, T. Non-leaching antibacterial cotton fabrics based on lipidated peptides. RSC Adv. 2017, 7, 34267–34275. [Google Scholar] [CrossRef] [Green Version]
- Sato, H.; Feix, J.B. Peptide–membrane interactions and mechanisms of membrane destruction by amphipathic α-helical antimicrobial peptides. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 1245–1256. [Google Scholar] [CrossRef]
- Lebl, M. Solid-phase synthesis on planar supports. Biopolym. (Pept. Sci.) 1998, 47, 397–404. [Google Scholar] [CrossRef]
- Tam, J.P. Synthetic peptide vaccine design: Synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 1988, 85, 5409–5413. [Google Scholar] [CrossRef]
- Tam, J.P.; Lu, Y.-A.; Yang, J.-L. Antimicrobial dendrimeric peptides. Eur. J. Biochem. 2002, 269, 923–932. [Google Scholar] [CrossRef]
- Kowalczyk, W.; Monsó, M.; de la Torre, B.G.; Andreu, D. Synthesis of multiple antigenic peptides (MAPs)—Strategies and limitations. J. Pept. Sci. 2011, 17, 247–251. [Google Scholar] [CrossRef]
- Raillard, S.P.; Mann, A.D.; Baer, T.A. An efficient procedure for the preparation of Fmoc-amino acids. Org. Prep. Proced. Int. 1998, 30, 183–186. [Google Scholar] [CrossRef]
- Kaiser, E.; Colescott, R.L.; Bossinger, C.D.; Cook, P.I. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal. Biochem. 1970, 34, 595–598. [Google Scholar] [CrossRef]
- ASTM E313-00 Standard Practice for Calculating Yellowness and Whiteness Indices from Instrumentally Measured Color Coordinates; ASTM International: West Conshohocken, PA, USA, 2000.
- Vallon-Eberhard, A.; Makovitzki, A.; Beauvais, A.; Latge, J.P.; Jung, S.; Shai, Y. Efficient clearance of Aspergillus fumigatus in murine lungs by an ultrashort antimicrobial lipopeptide, palmitoyl-Lys-Ala-DAla-Lys. Antimicrob. Agents Chemother. 2008, 52, 3118–3126. [Google Scholar] [CrossRef]
- Strøm, M.B.; Rekdal, Ø.; Svendsen, J.S. Antimicrobial activity of short arginine- and tryptophan-rich peptides. J. Pept. Sci. 2002, 8, 431–437. [Google Scholar] [CrossRef]
- Oh, D.; Shirazi, A.N.; Northup, K.; Sullivan, B.; Tiwari, R.K.; Bisoffi, M.; Parang, K. Enhanced cellular uptake of short polyarginine peptides through fatty acylation and cyclization. Mol. Pharmaceut. 2014, 11, 2845–2854. [Google Scholar] [CrossRef]
- Gholivand, K.; Dorosti, N. Novel ammonium phosphinates containing peptide moiety: Synthesis, structure, and in vitro antimicrobial activity. Chem. Pap. 2012, 66, 765–771. [Google Scholar] [CrossRef]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.-G.; Shai, Y. The Consequence of sequence alteration of an amphipathic α-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef]
- Doh, S.; Lee, J.; Lim, D.; Im, J. Manufacturing and analyses of wet-laid nonwoven consisting of carboxymethyl cellulose fibers. Fibers Polym. 2013, 14, 2176–2184. [Google Scholar] [CrossRef]
- Stevens, J.S.; de Luca, A.C.; Pelendritis, M.; Terenghi, G.; Downes, S.; Schroeder, S.L.M. Quantitative analysis of complex amino acids and RGD peptides by X-ray photoelectron spectroscopy (XPS). Surf. Interface Anal. 2013, 45, 1238–1246. [Google Scholar] [CrossRef] [Green Version]
- Palamutcu, S.; Keskin, R.; Sengul, M.; Devrent, N.; Ikis, Y.; Hascelik, B. Physical properties of antibacterial treated cotton fabrics and effect of laundry cycle. Ann. Univ. Oradea Fascicle Text. Leatherwork 2014, 15, 79–82. [Google Scholar]





| Sample | Peptide Loading (mmol/g) | IR (KBr) (cm−1) | |
|---|---|---|---|
| 1 | Pal-Lys-Ala-D-Ala-Lys-NH-CH2-CH2-N(Ac)-cotton | 0.76 | 1653, 1543 |
| 2 | Pal-His-Ala-D-Ala-His-NH-CH2-CH2-N(Ac)-cotton | 0.63 | 1643, 1540 |
| 3 | Pal-Arg-Ala-D-Ala-Arg-NH-CH2-CH2-N(Ac)-cotton | 0.60 1 | 1674, 1539, 1256 |
| 3′ | Pal-Arg(NO2)-Ala-D-Ala-Arg(NO2)-NH-CH2-CH2-N(Ac)-cotton | 0.60 | 1674, 1539, 1256 |
| 1a | [(Pal-Lys-Ala-D-Ala-Lys)4-Lys2]-Lys-NH-CH2-CH2-N(Ac)-cotton | 2.85 | 1734, 1654, 1532 |
| 2a | [(Pal-His-Ala-D-Ala-His)4-Lys2]-Lys- NH-CH2-CH2-N(Ac)-cotton | 2.12 | 1737, 1657, 1531 |
| 3a | [(Pal-Arg-Ala-D-Ala-Arg)4-Lys2]-Lys-NH-CH2-CH2-N(Ac)-cotton | 2.43 | 1733, 1653, 1534, 1249 |
| Sample | %C | %N | %O | |
|---|---|---|---|---|
| Cotton | 78.0 | 0.4 | 21.6 | |
| Cotton-N(Ac)-CH2-CH2-NH2 | 59.8 | 1.7 | 38.5 | |
| 1 | Pal-Lys-Ala-D-Ala-Lys-NH-CH2-CH2-N(Ac)-Cotton | 68.1 | 4.7 | 27.2 |
| 2 | Pal-His-Ala-D-Ala-His-NH-CH2-CH2-N(Ac)-Cotton | 71.4 | 4.9 | 23.7 |
| 3 | Pal-Arg-Ala-D-Ala-Arg-NH-CH2-CH2-N(Ac)-Cotton | 69.2 | 4.8 | 26.0 |
| 3′ | Pal-Arg(NO2)-Ala-D-Ala-Arg(NO2)-NH-CH2-CH2-N(Ac)-cotton | 72.6 | 6.3 | 21.1 |
| 1a | [(Pal-Lys-Ala-D-Ala-Lys)4-Lys2]-Lys-NH-CH2-CH2-N(Ac)-cotton | 70.3 | 8.9 | 20.8 |
| 2a | [(Pal-His-Ala-D-Ala-His)4-Lys2]-Lys-NH-CH2-CH2-N(Ac)-cotton | 71.2 | 7.3 | 22.5 |
| 3a | [(Pal-Arg-Ala-D-Ala-Arg)4-Lys2]-Lys-NH-CH2-CH2-N(Ac)-cotton | 70.0 | 8.1 | 21.9 |
| Sample | Side Chain | Nitrogen Species | Binding Energy (eV) | Area (%) |
|---|---|---|---|---|
| 1 |  | Amide | 400.6 | 28.6 |
| Amine | 401.5 | 34.1 | ||
| Alkyl ammonium | 401.8 | 37.3 | ||
| 2 |  | Imino imidazole | 398.3 | 11.0 |
| Amino imidazole | 399.5 | 9.2 | ||
| Amide | 400.9 | 77.7 | ||
| Protonated amide | 403.4 | 2.1 | ||
| 3′ |  | Imino guanidinium | 398.4 | 3.0 |
| Amino guanidinium | 399.8 | 24.2 | ||
| Amide | 400.9 | 43.2 | ||
| Amine | 401.7 | 25.3 | ||
| Nitro | 402.8 | 4.3 |
| Sample | Berger Whiteness | CIE Whiteness Index | Yellowness Index |
|---|---|---|---|
| Cotton | 60.84 | 62.45 | 7.59 |
| 1 | 49.67 | 50.0 | 11.28 |
| 2 | 50.29 | 51.5 | 9.87 |
| 3 | 25.35 | 26.2 | 19.54 |
| 2a | 57.96 | 57.37 | 8.16 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orlandin, A.; Dolcet, P.; Biondi, B.; Hilma, G.; Coman, D.; Oancea, S.; Formaggio, F.; Peggion, C. Covalent Graft of Lipopeptides and Peptide Dendrimers to Cellulose Fibers. Coatings 2019, 9, 606. https://doi.org/10.3390/coatings9100606
Orlandin A, Dolcet P, Biondi B, Hilma G, Coman D, Oancea S, Formaggio F, Peggion C. Covalent Graft of Lipopeptides and Peptide Dendrimers to Cellulose Fibers. Coatings. 2019; 9(10):606. https://doi.org/10.3390/coatings9100606
Chicago/Turabian StyleOrlandin, Andrea, Paolo Dolcet, Barbara Biondi, Geta Hilma, Diana Coman, Simona Oancea, Fernando Formaggio, and Cristina Peggion. 2019. "Covalent Graft of Lipopeptides and Peptide Dendrimers to Cellulose Fibers" Coatings 9, no. 10: 606. https://doi.org/10.3390/coatings9100606
APA StyleOrlandin, A., Dolcet, P., Biondi, B., Hilma, G., Coman, D., Oancea, S., Formaggio, F., & Peggion, C. (2019). Covalent Graft of Lipopeptides and Peptide Dendrimers to Cellulose Fibers. Coatings, 9(10), 606. https://doi.org/10.3390/coatings9100606