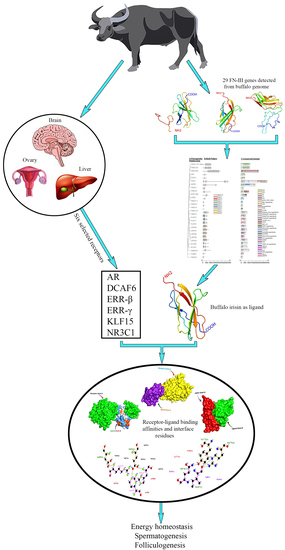
Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of FN-III Family Genes and Characterization of Their Physiochemical Properties
2.2. Collinearity and Multiple Sequence Alignment Analysis
2.3. Molecular Docking Analysis
3. Results
3.1. Identification of FN-III Gene Family and Their Physiochemical Properties
3.2. Gene Structure and Motif Analysis
3.3. Collinearity Analysis of FN-III Gene Family
3.4. Structural Configuration of FNDC5 Protein
3.5. Multiple Sequence Alignment Analysis of Irisin
3.6. Molecular Docking Analysis of FNDC5/Irisin
4. Discussion
4.1. The Identification and Characterization of FN-III Gene Family
4.2. Structural Configuration of FNDC5 Protein and Its Potential Role in Cellular Activities
4.3. Molecular Docking Analysis of FNDC5/Irisin with Selected Receptors
4.3.1. FNDC5/Irisin and Estrogen-Related Receptors
4.3.2. FNDC5/Irisin and Androgen Receptors
4.3.3. FNDC5/Irisin and Glucocorticoid Receptors
4.3.4. FNDC5/Irisin and Krüppel-like Factor 15 Receptors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pasha, T.; Hayat, Z. Present situation and future perspective of buffalo production in Asia. J. Anim. Plant Sci. 2012, 22, 250–256. [Google Scholar]
- Rehman, S.U.; Shafique, L.; Yousuf, M.R.; Liu, Q.; Ahmed, J.Z.; Riaz, H. Spectrophotometric Calibration and Comparison of Different Semen Evaluation Methods in Nili-Ravi Buffalo Bulls. Pak. Vet. J. 2019, 39, 568–572. [Google Scholar] [CrossRef]
- Imran, S.; Javed, M.; Yaqub, T.; Iqbal, M.; Nadeem, A.; Mukhtar, N.; Maccee, F. Genetic basis of estrous in bovine: A Review. Int. J. Adv. Res. 2014, 2, 962–966. [Google Scholar]
- Li, Z.; Lu, S.; Cui, K.; Shafique, L.; ur Rehman, S.; Luo, C.; Wang, Z.; Ruan, J.; Qian, Q.; Liu, Q. Fatty acid biosynthesis and transcriptional regulation of Stearoyl-CoA Desaturase 1 (SCD1) in buffalo milk. BMC Genet. 2020, 21, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehman, S.u.; Nadeem, A.; Javed, M.; Hassan, F.U.; Luo, X.; Khalid, R.B.; Liu, Q. Genomic Identification, Evolution and Sequence Analysis of the Heat-Shock Protein Gene Family in Buffalo. Genes 2020, 11, 1388. [Google Scholar] [CrossRef]
- Rehman, S.u.; Feng, T.; Wu, S.; Luo, X.; Lei, A.; Luobu, B.; Hassan, F.-U.; Liu, Q. Comparative Genomics, Evolutionary and Gene Regulatory Regions Analysis of Casein Gene Family in Bubalus bubalis. Front. Genet. 2021, 12, 408. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.; Hassan, F.-u.; Luo, X.; Li, Z.; Liu, Q. Whole-Genome Sequencing and Characterization of Buffalo Genetic Resources: Recent Advances and Future Challenges. Animals 2021, 11, 904. [Google Scholar] [CrossRef]
- Shi, W.; Yuan, X.; Cui, K.; Li, H.; Fu, P.; Rehman, S.-U.; Shi, D.; Liu, Q.; Li, Z. LC-MS/MS Based Metabolomics Reveal Candidate Biomarkers and Metabolic Changes in Different Buffalo Species. Animals 2021, 11, 560. [Google Scholar] [CrossRef] [PubMed]
- De Rensis, F.; Lopez-Gatius, F. Protocols for synchronizing estrus and ovulation in buffalo (Bubalus bubalis): A review. Theriogenology 2007, 67, 209–216. [Google Scholar] [CrossRef]
- Crowe, M.A.; Mullen, M.P. Regulation and Function of Gonadotropins throughout the Bovine Oestrous Cycle. In Crowe and Mullen; InTech: London, UK, 2013; Volume 143. [Google Scholar]
- Hassan, F.; Khan, M.; Rehman, M.; Sarwar, M.; Bhatti, S. Seasonality of calving in Nili-Ravi buffaloes, purebred Sahiwal and crossbred cattle in Pakistan. Ital. J. Anim. Sci. 2007, 6, 1298–1301. [Google Scholar] [CrossRef]
- Perera, B. Reproductive cycles of buffalo. Anim. Reprod. Sci. 2011, 124, 194–199. [Google Scholar] [CrossRef]
- Hussain Shah, S.N. Prolonged calving intervals in the Nili Ravi buffalo. Ital. J. Anim. Sci. 2007, 6, 694–696. [Google Scholar] [CrossRef]
- Warriach, H.; McGill, D.; Bush, R.; Wynn, P.; Chohan, K. A review of recent developments in buffalo reproduction—A review. Asian-Australasian. J. Anim. Sci. 2015, 28, 451. [Google Scholar]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenick, L.V.; Hsia, H.C.; Schwarzbauer, J.E. Fibronectin fragmentation promotes α4β1 integrin-mediated contraction of a fibrin–fibronectin provisional matrix. Exp. Cell Res. 2005, 309, 48–55. [Google Scholar] [CrossRef]
- Wagenmakers, A.J.; Hawley, J.A.; Hargreaves, M.; Zierath, J.R. Signalling mechanisms in skeletal muscle: Role in substrate selection and muscle adaptation. Essays Biochem. 2006, 42, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Timmons, J.A.; Baar, K.; Davidsen, P.K.; Atherton, P.J. Is irisin a human exercise gene? Nature 2012, 488, E9–E10. [Google Scholar] [CrossRef]
- Perakakis, N.; Triantafyllou, G.A.; Fernández-Real, J.M.; Huh, J.Y.; Park, K.H.; Seufert, J.; Mantzoros, C.S. Physiology and role of irisin in glucose homeostasis. Nat. Rev. Endocrinol. 2017, 13, 324–337. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Navarrete, J.M.; Ortega, F.; Serrano, M.; Guerra, E.; Pardo, G.; Tinahones, F.; Ricart, W.; Fernández-Real, J.M. Irisin is expressed and produced by human muscle and adipose tissue in association with obesity and insulin resistance. J. Clin. Endocrinol. Metab. 2013, 98, E769–E778. [Google Scholar] [CrossRef] [PubMed]
- Roca-Rivada, A.; Castelao, C.; Senin, L.L.; Landrove, M.O.; Baltar, J.; Crujeiras, A.B.; Seoane, L.M.; Casanueva, F.F.; Pardo, M. FNDC5/irisin is not only a myokine but also an adipokine. PLoS ONE 2013, 8, e60563. [Google Scholar]
- Aydin, S.; Kuloglu, T.; Aydin, S.; Kalayci, M.; Yilmaz, M.; Cakmak, T.; Albayrak, S.; Gungor, S.; Colakoglu, N.; Ozercan, İ.H. A comprehensive immunohistochemical examination of the distribution of the fat-burning protein irisin in biological tissues. Peptides 2014, 61, 130–136. [Google Scholar] [CrossRef] [PubMed]
- NCBI. 2021. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 3 March 2021).
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Bank, P.D. Protein Data Bank; Springer: Dordrecht, The Netherlands, 1971. [Google Scholar]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Plopper, G. The Extracellular Matrix and Cell Adhesion in CELLS; Lewin, B., Cassimeris, L., Lingappa, V., Plopper, G., Eds.; Jones and Bartlett: Sudbury, MA, USA, 2007; pp. 645–702. ISBN 978-0-7637-3905-8. [Google Scholar]
- Williams, C.M.; Engler, A.J.; Slone, R.D.; Galante, L.L.; Schwarzbauer, J.E. Fibronectin expression modulates mammary epithelial cell proliferation during acinar differentiation. Cancer Res. 2008, 68, 3185–3192. [Google Scholar] [CrossRef] [Green Version]
- Tominaga, K.; Johmura, Y.; Nishizuka, M.; Imagawa, M. Fad24, a mammalian homolog of Noc3p, is a positive regulator in adipocyte differentiation. J. Cell Sci. 2004, 117, 6217–6226. [Google Scholar] [CrossRef] [Green Version]
- Raschke, S.; Elsen, M.; Gassenhuber, H.; Sommerfeld, M.; Schwahn, U.; Brockmann, B.; Jung, R.; Wisløff, U.; Tjønna, A.E.; Raastad, T. Evidence against a beneficial effect of irisin in humans. PLoS ONE 2013, 8, e73680. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Song, W.; Hu, T.; Zhang, N.; Miao, S.; Zong, S.; Wang, L. Fank1 interacts with Jab1 and regulates cell apoptosis via the AP-1 pathway. Cell. Mol. Sci. 2011, 68, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhang, X.; Li, M.; Ma, X.; Huang, B.; Chen, H.; Chen, D. Proapoptotic RYBP interacts with FANK1 and induces tumor cell apoptosis through the AP-1 signaling pathway. Cell. Signal. 2016, 28, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Manabe, R.-I.; Whitmore, L.; Weiss, J.M.; Horwitz, A.R. Identification of a novel microtubule-associated protein that regulates microtubule organization and cytokinesis by using a GFP-screening strategy. Curr. Biol. 2002, 12, 1946–1951. [Google Scholar] [CrossRef] [Green Version]
- Stein, P.A.; Toret, C.P.; Salic, A.N.; Rolls, M.M.; Rapoport, T.A. A novel centrosome-associated protein with affinity for microtubules. J. Cell Sci. 2002, 115, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Seabold, G.K.; Wang, P.Y.; Chang, K.; Wang, C.-Y.; Wang, Y.-X.; Petralia, R.S.; Wenthold, R.J. The SALM family of adhesion-like molecules forms heteromeric and homomeric complexes. J. Biol. Chem. 2008, 283, 8395–8405. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.Y.; Seabold, G.K.; Wenthold, R.J. Synaptic adhesion-like molecules (SALMs) promote neurite outgrowth. Mol. Cell. Neurosci. 2008, 39, 83–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsuji, L.; Yamashita, T.; Kubo, T.; Madura, T.; Tanaka, H.; Hosokawa, K.; Tohyama, M. FLRT3, a cell surface molecule containing LRR repeats and a FNIII domain, promotes neurite outgrowth. Biochem. Biophys. Res. Commun. 2004, 313, 1086–1091. [Google Scholar] [CrossRef]
- Lu, Y.C.; Nazarko, O.V.; Sando III, R.; Salzman, G.S.; Li, N.-S.; Südhof, T.C.; Araç, D. Structural basis of latrophilin-FLRT-UNC5 interaction in cell adhesion. Structure 2015, 23, 1678–1691. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Yun, S.; Choi, G.H.; Jung, T.W. Fibronectin type III domain containing 4 attenuates hyperlipidemia-induced insulin resistance via suppression of inflammation and ER stress through HO-1 expression in adipocytes. Biochem. Biophys. Res. Commun. 2018, 502, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; Van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009, 16, 365–371. [Google Scholar] [CrossRef] [Green Version]
- Obholz, K.L.; Akopyan, A.; Waymire, K.G.; MacGregor, G.R. FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev. Biol. 2006, 298, 498–513. [Google Scholar] [CrossRef] [Green Version]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Ferrer-Martínez, A.; Ruiz-Lozano, P.; Chien, K.R. Mouse PeP: A novel peroxisomal protein linked to myoblast differentiation and development. Dev. Dyn. 2002, 224, 154–167. [Google Scholar] [CrossRef]
- Teufel, A.; Malik, N.; Mukhopadhyay, M.; Westphal, H. Frcp1 and Frcp2, two novel fibronectin type III repeat containing genes. Gene 2002, 297, 79–83. [Google Scholar] [CrossRef]
- Aydin, S. Three new players in energy regulation: Preptin, adropin and irisin. Peptides 2014, 56, 94–110. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Chinnam, N.; Ohashi, T.; Shah, R.S.; Erickson, H.P. The structure of irisin reveals a novel intersubunit β-sheet fibronectin type III (FNIII) dimer: Implications for receptor activation. J. Biol. Chem. 2013, 288, 33738–33744. [Google Scholar] [CrossRef] [Green Version]
- Wahab, F.; Khan, I.U.; Polo, I.R.; Zubair, H.; Drummer, C.; Shahab, M.; Behr, R. Irisin in the primate hypothalamus and its effect on GnRH in vitro. J. Endocrinol. 2019, 241, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Wahab, F.; Drummer, C.; Mätz-Rensing, K.; Fuchs, E.; Behr, R. Irisin is expressed by undifferentiated spermatogonia and modulates gene expression in organotypic primate testis cultures. Mol. Cell. Endocrinol. 2020, 504, 110670. [Google Scholar] [CrossRef]
- Shi, H.; Kumar, S.P.D.S.; Liu, X. G protein-coupled estrogen receptor in energy homeostasis and obesity pathogenesis. In Progress in Molecular Biology and Translational Science; Elsevier: Oxford, UK, 2013; Volume 114, pp. 193–250. [Google Scholar]
- Pathak, D.; Bansal, N.; Singh, O.; Gupta, K.; Ghuman, S. Immunohistochemical localization of estrogen receptor alpha (ERα) in the oviduct of Indian buffalo during follicular and luteal phases of estrous cycle. In Tropical Animal Health and Production; Springer Nature: Cham, Switzerland, 2019; Volume 51, pp. 1601–1609. [Google Scholar]
- Saruhan, B.; Saǧsoz, H.; Akbalik, M.; Ketani, M. Distribution of estrogen receptor α and progesterone receptor B in the bovine oviduct during the follicular and luteal phases of the sexual cycle: An immunohistochemical and semi-quantitative study. Biotech. Histochem. 2011, 86, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Iavarone, M.; Lampertico, P.; Seletti, C.; Francesca Donato, M.; Ronchi, G.; Del Ninno, E.; Colombo, M. The clinical and pathogenetic significance of estrogen receptor-β expression in chronic liver diseases and liver carcinoma. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2003, 98, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Miceli, V.; Cocciadiferro, L.; Fregapane, M.; Zarcone, M.; Montalto, G.; Polito, L.M.; Agostara, B.; Granata, O.M.; Carruba, G. Expression of wild-type and variant estrogen receptor alpha in liver carcinogenesis and tumor progression. Omics A J. Integr. Biol. 2011, 15, 313–317. [Google Scholar] [CrossRef]
- Cui, J.; Shen, Y.; Li, R. Estrogen synthesis and signaling pathways during aging: From periphery to brain. Trends Mol. Med. 2013, 19, 197–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuiper, G.G.; Carlsson, B.; Grandien, K.; Enmark, E.; Häggblad, J.; Nilsson, S.; Gustafsson, J.-A.k. Comparison of the ligand binding specificity and transcript tissue distribution of estrogen receptors α and β. Endocrinology 1997, 138, 863–870. [Google Scholar] [CrossRef]
- Alvaro, D.; Alpini, G.; Onori, P.; Perego, L.; Baroni, G.S.; Franchitto, A.; Baiocchi, L.; Glaser, S.S.; Le Sage, G.; Folli, F. Estrogens stimulate proliferation of intrahepatic biliary epithelium in rats. Gastroenterology 2000, 119, 1681–1691. [Google Scholar] [CrossRef]
- Heard, D.J.; Norby, P.L.; Holloway, J.; Vissing, H. Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: Tissue-specific isoforms are expressed during development and in the adult. Mol. Endocrinol. 2000, 14, 382–392. [Google Scholar] [CrossRef] [Green Version]
- Sanyal, S.; Matthews, J.; Bouton, D.; Kim, H.-J.; Choi, H.-S.; Treuter, E.; Gustafsson, J.-A.k. Deoxyribonucleic acid response element-dependent regulation of transcription by orphan nuclear receptor estrogen receptor-related receptor γ. Mol. Endocrinol. 2004, 18, 312–325. [Google Scholar] [CrossRef] [Green Version]
- Powelka, A.M.; Seth, A.; Virbasius, J.V.; Kiskinis, E.; Nicoloro, S.M.; Guilherme, A.; Tang, X.; Straubhaar, J.; Cherniack, A.D.; Parker, M.G. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J. Clin. Investig. 2006, 116, 125–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamei, Y.; Ohizumi, H.; Fujitani, Y.; Nemoto, T.; Tanaka, T.; Takahashi, N.; Kawada, T.; Miyoshi, M.; Ezaki, O.; Kakizuka, A. PPARγ coactivator 1β/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. USA 2003, 100, 12378–12383. [Google Scholar] [CrossRef] [Green Version]
- Varela-Rodríguez, B.M.; Pena-Bello, L.; Juiz-Valiña, P.; Vidal-Bretal, B.; Cordido, F.; Sangiao-Alvarellos, S. FNDC5 expression and circulating irisin levels are modified by diet and hormonal conditions in hypothalamus, adipose tissue and muscle. Sci. Rep. 2016, 6, 29898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, J.Y.; Panagiotou, G.; Mougios, V.; Brinkoetter, M.; Vamvini, M.T.; Schneider, B.E.; Mantzoros, C.S. FNDC5 and irisin in humans: I. Predictors of circulating concentrations in serum and plasma and II. mRNA expression and circulating concentrations in response to weight loss and exercise. Metabolism 2012, 61, 1725–1738. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhang, X.-Y.; Sun, Y.; Hou, X.-G.; Chen, L. Higher circulating irisin levels in patients with polycystic ovary syndrome: A meta-analysis. Gynecol. Endocrinol. 2018, 34, 290–293. [Google Scholar] [CrossRef]
- Salem, H.; Yatchenko, Y.; Anosov, M.; Rosenfeld, T.; Altarescu, G.; Grisaru-Granovsky, S.; Birk, R. Maternal and neonatal irisin precursor gene FNDC5 polymorphism is associated with preterm birth. Gene 2018, 649, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Basini, G.; Bussolati, S.; Iannarelli, M.; Ragionieri, L.; Grolli, S.; Ramoni, R.; Dodi, A.; Gazza, F.; Grasselli, F. The myokine irisin: Localization and effects in swine late medium and large antral ovarian follicle. Domest. Anim. Endocrinol. 2021, 74, 106576. [Google Scholar] [CrossRef]
- Hasbi, H.; Gustina, S. Androgen Regulation in Spermatogenesis to Increase Male Fertility. WARTAZOA. Indones. Bull. Anim. Vet. Sci. 2018, 28, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Tsai, T.-C.; Lee, Y.-L.; Hsiao, W.-C.; Tsao, Y.-P.; Chen, S.-L. NRIP, a novel nuclear receptor interaction protein, enhances the transcriptional activity of nuclear receptors. J. Biol. Chem. 2005, 280, 20000–20009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular architecture and assembly of the DDB1–CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef]
- Rhen, T.; Cidlowski, J.A. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N. Engl. J. Med. 2005, 353, 1711–1723. [Google Scholar] [CrossRef] [Green Version]
- Lu, N.Z.; Wardell, S.E.; Burnstein, K.L.; Defranco, D.; Fuller, P.J.; Giguere, V.; Hochberg, R.B.; McKay, L.; Renoir, J.-M.; Weigel, N.L. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: Glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol. Rev. 2006, 58, 782–797. [Google Scholar] [CrossRef] [PubMed]
- Pratt, W.; Morishima, Y.; Murphy, M.; Harrell, M. Chaperoning of glucocorticoid receptors. In Molecular Chaperones in Health and Disease; Springer: Berlin/Heidelberg, Germany, 2006; pp. 111–138. [Google Scholar]
- Buckingham, J.C. Glucocorticoids: Exemplars of multi-tasking. Br. J. Pharmacol. 2006, 147, S258–S268. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, R.; Wada, H.; Ito, K.; Adcock, I.M. Effects of glucocorticoids on gene transcription. Eur. J. Pharmacol. 2004, 500, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Ray, A.; Prefontaine, K.E. Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 752–756. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, E.P.; Nader, G.A. Balancing muscle hypertrophy and atrophy. Nat. Med. 2004, 10, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.; Wang, B.; Orihuela, Y.; Hong, E.-G.; Fisch, S.; Haldar, S.; Cline, G.W.; Kim, J.K.; Peroni, O.D.; Kahn, B.B. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007, 5, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, N.; Yoshikawa, N.; Ito, N.; Maruyama, T.; Suzuki, Y.; Takeda, S.-I.; Nakae, J.; Tagata, Y.; Nishitani, S.; Takehana, K. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011, 13, 170–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene | Chr. | Exon Count | MW (Da) | A.A | pI | AI | II | GRAVY |
|---|---|---|---|---|---|---|---|---|
| Fibronectin 1 (FN1) | 2 | 46 | 258,641.53 | 2354 | 5.28 | 69.74 | 40.09 | −0.487 |
| Fibronectin type III domain containing 5 (FNDC5) | 2 | 6 | 22,869.33 | 205 | 6.44 | 92.68 | 52.30 | −0.218 |
| Fibronectin type III domain containing 3B (FNDC3B) | 1 | 31 | 127,736.34 | 1160 | 5.91 | 69.91 | 53.98 | −0.434 |
| Fibronectin type III and ankyrin repeat domains 1 (FANK1) | 23 | 14 | 38,413.93 | 345 | 8.51 | 89.51 | 33.76 | −0.334 |
| Fibronectin type III and SPRY domain containing 1 like (FSD1L) | 3 | 16 | 58,607.09 | 521 | 6.32 | 75.93 | 46.15 | −0.574 |
| Leucine-rich repeat and fibronectin type III domain containing 1 (LRFN1) | 18 | 8 | 82,023.66 | 770 | 7.89 | 90.16 | 49.73 | −0.066 |
| Leucine-rich repeat and fibronectin type III domain containing 5 (LRFN5) | 20 | 8 | 52,122.68 | 466 | 6.60 | 95.47 | 35.44 | −0.141 |
| Fibronectin type III and SPRY domain containing 1 (FSD1) | 9 | 13 | 55,768.58 | 662 | 4.96 | 77.88 | 48.72 | −0.380 |
| Fibronectin type III domain containing 3A (FNDC3A) | 13 | 31 | 133,632.56 | 1217 | 6.44 | 71.27 | 46.88 | −0.412 |
| Fibronectin type III domain containing 1 (FNDC1) | 10 | 23 | 205,865.78 | 1905 | 9.66 | 59.01 | 59.92 | −0.799 |
| Leucine-rich repeat and fibronectin type III domain containing 3 (LRFN3) | 18 | 5 | 72,450.76 | 679 | 9.38 | 87.05 | 59.78 | −0.246 |
| Fibronectin type III and SPRY domain containing 2 (FSD2) | 20 | 15 | 84,755.73 | 747 | 4.81 | 69.69 | 47.20 | −0.593 |
| Fibronectin type III domain containing 7 (FNDC7) | 6 | 13 | 85,949.11 | 811 | 6.53 | 77.69 | 45.18 | 0.046 |
| Ankyrin repeat and fibronectin type III domain containing 1 (ANKFN1) | 3 | 20 | 120,567.79 | 1068 | 6.52 | 80.73 | 58.15 | −0.467 |
| Immunoglobulin like and fibronectin type III domain containing 1 (IGFN1) | 5 | 26 | 347,525.99 | 3490 | 6.49 | 55.35 | 34.98 | −0.590 |
| Fibronectin type III domain containing 4 (FNDC4) | 12 | 7 | 24,753.16 | 230 | 7.66 | 88.87 | 55.08 | −0.252 |
| Fibronectin type III domain containing 8 (FNDC8) | 3 | 4 | 34,298.93 | 312 | 5.29 | 80.93 | 46.44 | −0.370 |
| Leucine-rich repeat and fibronectin type III domain containing 4 (LRFN4) | 5 | 3 | 66,839.10 | 636 | 6.70 | 94.14 | 42.55 | −0.028 |
| Fibronectin type III domain containing protein 3C1-like (LOC102393884) | X | 27 | 157,320.54 | 1433 | 6.79 | 71.84 | 45.92 | −0.439 |
| Fibronectin leucine-rich transmembrane protein 2 (FLRT2) | 11 | 4 | 73,773.40 | 660 | 7.89 | 94.18 | 36.58 | −0.185 |
| EGF like, fibronectin type III and laminin G domains (EGFLAM) | 19 | 23 | 112,751.54 | 1032 | 6.53 | 74.46 | 41.70 | −0.325 |
| Fibronectin type III domain containing 9 (FNDC9) | 9 | 2 | 25,342.98 | 227 | 5.65 | 85.99 | 54.56 | −0.055 |
| Leucine-rich repeat and fibronectin type III domain containing 2 (LRFN2) | 2 | 2 | 87,694.08 | 820 | 6.59 | 90.88 | 44.73 | −0.097 |
| Fibronectin leucine-rich transmembrane protein 3 (FLRT3) | 14 | 3 | 73,171.75 | 649 | 7.56 | 94.18 | 44.53 | −0.296 |
| Fibronectin leucine-rich transmembrane protein 1 (FLRT1) | 5 | 2 | 74,144.68 | 677 | 6.15 | 96.88 | 32.12 | −0.122 |
| Fibronectin type III domain containing 11 (FNDC11) | 14 | 4 | 38,198.37 | 333 | 6.81 | 96.34 | 53.23 | −0.280 |
| Fibronectin type III domain containing 10 (FNDC10) | 5 | 3 | 24,097.32 | 225 | 9.11 | 87.33 | 66.15 | 0.124 |
| Extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ELFN2) | 4 | 4 | 90,363.67 | 824 | 7.78 | 81.78 | 48.76 | −0.295 |
| Extracellular leucine-rich repeat and fibronectin type III domain containing 1 (ELFN1) | 24 | 3 | 87,687.60 | 808 | 8.82 | 79.43 | 61.89 | −0.351 |
| Motif | Protein Sequence | Length | Pfam Domain |
|---|---|---|---|
| MEME-1 | DNFIAAIPRRDFANMTGLVDLTLSRNTISHIEAGAFDDLENLRALHLDNN | 50 | LRR_8 |
| MEME-2 | NPLHCNCELLWLRRLAREDDLETCASPPGLTGRYFWSVPEEEFLCEPPLI | 50 | LRRCT |
| MEME-3 | LTNLEPDTTYRLCVQALNSAG | 21 | fn3 |
| MEME-4 | MVNLETLRLDHNLIDTIPPGAFSELHKLARLDLTSNRLQKL | 41 | LRR_8 |
| MEME-5 | HWVAPDGRLVGNSSRTRVYPNGTLDILITTSGDSGAFTCIASNAAGEATA | 50 | I-set |
| MEME-6 | CPSVCRCDRGFIYCNDRGLTSIPAGIPEDATTLYLQNNQINNAGIPADLK | 50 | LRRNT |
| MEME-7 | CPKRCICQNLSPSLSTLCAKKGLLFVPPNIDRRTVELRL | 39 | Toxin_11 |
| MEME-8 | WPVQRPAPGIRMYQIQYNSSADDTLVYRM | 29 | - |
| MEME-9 | LEDLDLSYNNLESIPW | 16 | LRR_4 |
| MEME-10 | GTEYRFRVRACNEAGEGPLSEPYTVTTPP | 29 | fn3 |
| Sr. No. | Receptor | Docking Score | Ligand RMSD (A0) | Ligand Interacting Residues |
|---|---|---|---|---|
| 1 | Androgen | −311.40 | 86.42 | Asn36, Thr38, Arg40 |
| 2 | DDB1 and CUL4 associated factor 6 | −256.63 | 79.76 | Asn36, Thr38, Arg40, His41 |
| 3 | Estrogen-related receptor β | −295.57 | 108.96 | Arg72, Mse73, Leu74, Arg75, Phe76, Ile77, Gln78, Glu79, Val80, Asn81, Cys87, Ala88, Trp90, Asp91 |
| 4 | Estrogen-related receptor γ | −256.63 | 79.76 | Arg40, His41, Lys43, Lys120, Pro122, Arg123 |
| 5 | Krüppel-like factor 15 | −260.71 | 81.85 | Ser30, Pro31, Arg72, Mse73, Leu74, Arg75, Phe76, Ile77, Gln78, Glu79, Asn81, Ala88, Trp90, Gln108, Pro112, Val180 |
| 6 | Nuclear receptor subfamily 3 group C member 1 | −308.59 | 108.34 | Lys740, Glu741, Asn742, Leu744, Leu745, Arg746, Leu748, Leu749, Asp753 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, S.; Hassan, F.-u.; Luo, Y.; Fatima, I.; Ahmed, I.; Ihsan, A.; Safdar, W.; Liu, Q.; Rehman, S.u. Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors. Biology 2021, 10, 1207. https://doi.org/10.3390/biology10111207
Wu S, Hassan F-u, Luo Y, Fatima I, Ahmed I, Ihsan A, Safdar W, Liu Q, Rehman Su. Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors. Biology. 2021; 10(11):1207. https://doi.org/10.3390/biology10111207
Chicago/Turabian StyleWu, Siwen, Faiz-ul Hassan, Yuhong Luo, Israr Fatima, Ishtiaq Ahmed, Awais Ihsan, Warda Safdar, Qingyou Liu, and Saif ur Rehman. 2021. "Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors" Biology 10, no. 11: 1207. https://doi.org/10.3390/biology10111207
APA StyleWu, S., Hassan, F. -u., Luo, Y., Fatima, I., Ahmed, I., Ihsan, A., Safdar, W., Liu, Q., & Rehman, S. u. (2021). Comparative Genomic Characterization of Buffalo Fibronectin Type III Domain Proteins: Exploring the Novel Role of FNDC5/Irisin as a Ligand of Gonadal Receptors. Biology, 10(11), 1207. https://doi.org/10.3390/biology10111207