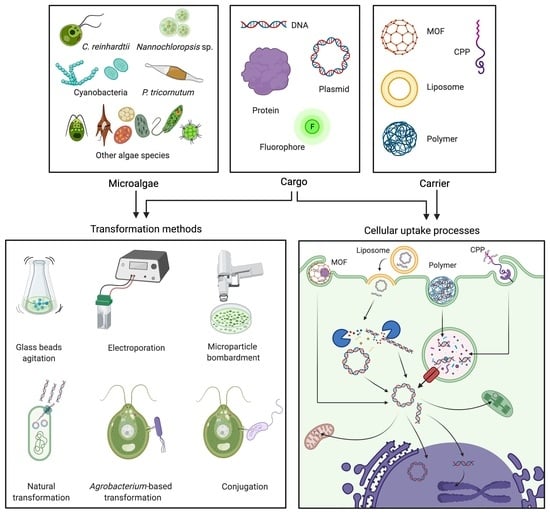
Gene Delivery Technologies with Applications in Microalgal Genetic Engineering
Abstract
:Simple Summary
Abstract
1. Introduction
2. Traditional Algal Transformation Techniques
2.1. Agitation of Cells in the Presence of DNA and Non-Ionic Surfactants
2.2. Electroporation
2.3. Microparticle Bombardment
3. Natural Transformation, Bacterial Conjugation, and Agrobacterium-Mediated Transformation
3.1. Natural Transformation
3.2. Bacterial Conjugation
3.3. Agrobacterium-Mediated Transformation
4. Non-Traditional and Emerging Transformation Technologies
4.1. Cell-Penetrating Peptides
4.2. Cell-Penetrating Polymers
4.3. Metal-Organic Frameworks
4.4. Liposome-Mediated Transformation
5. Considerations for the Future of Algal Transformation
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hallmann, A. Algal Transgenics and Biotechnology. Transgenic Plant J. 2007, 1, 81–98. [Google Scholar]
- Fabris, M.; Abbriano, R.M.; Pernice, M.; Sutherland, D.L.; Commault, A.S.; Hall, C.C.; Labeeuw, L.; Mccauley, J.I.; Kuzhiuparambil, U.; Ray, P.; et al. Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 2020, 11, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Nelson, D.R.; Hazzouri, K.M.; Lauersen, K.J.; Jaiswal, A.; Chaiboonchoe, A.; Mystikou, A.; Fu, W.; Daakour, S.; Dohai, B.; Alzahmi, A.; et al. Large-scale genome sequencing reveals the driving forces of viruses in microalgal evolution. Cell Host Microbe 2021, 29, 250–266.e8. [Google Scholar] [CrossRef]
- Benedetti, M.; Vecchi, V.; Barera, S.; Dall’Osto, L. Biomass from microalgae: The potential of domestication towards sustainable biofactories. Microb. Cell Factories 2018, 17, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factories 2018, 17, 1–21. [Google Scholar] [CrossRef]
- Chen, H.; Li, T.; Wang, Q. Ten years of algal biofuel and bioproducts: Gains and pains. Planta 2019, 249, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Chaiboonchoe, A.; Khraiwesh, B.; Nelson, D.R.; Al-Khairy, D.; Mystikou, A.; Alzahmi, A.; Salehi-Ashtiani, K. Algal cell factories: Approaches, applications, and potentials. Mar. Drugs 2016, 14, 225. [Google Scholar] [CrossRef] [Green Version]
- Wannathong, T.; Waterhouse, J.C.; Young, R.E.B.; Economou, C.K.; Purton, S. New tools for chloroplast genetic engineering allow the synthesis of human growth hormone in the green alga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2016, 100, 5467–5477. [Google Scholar] [CrossRef] [Green Version]
- Coll, J.M. Review. Methodologies for transferring DNA into eukaryotic microalgae. Spanish J. Agric. Res. 2006, 4, 316–330. [Google Scholar] [CrossRef] [Green Version]
- Porter, R.D. Transformation in cyanobacteria. Crit. Rev. Microbiol. 1986, 13, 111–132. [Google Scholar] [CrossRef] [PubMed]
- León, R.; Fernández, E.; Leon, R.; Fernandez, E. Nuclear transformation of eukaryotic microalgae: Historical overview, achievements and problems. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2007; Volume 616, pp. 1–11. ISBN 9780387755311. [Google Scholar]
- Klein, R.M.; Wolf, E.D.; Wu, R.; Sanford, J.C. High-velocity microprojectiles for delivering nucleic acids into living cells. 1987. Biotechnology 1992, 24, 384–386. [Google Scholar] [PubMed]
- Kindle, K.L.; Schnell, R.A.; Fernandez, E.; Lefebvre, P.A. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J. Cell Biol. 1989, 109, 2589–2601. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef] [Green Version]
- Sodeinde, O.A.; Kindle, K.L. Homologous recombination in the nuclear genome of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1993, 90, 9199–9203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.C.; Long, J.R.; Yaghi, O.M. Introduction to metal-organic frameworks. Chem. Rev. 2012, 112, 673–674. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Almeida, A.J.; Vale, N. Combination of cell-penetrating peptides with nanoparticles for therapeutic application: A review. Biomolecules 2019, 9, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanigan, T.M.; Kopera, H.C.; Saunders, T.L. Principles of Genetic Engineering. Genes 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hawkins, R.L.; Nakamura, M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr. Microbiol. 1999, 38, 335–341. [Google Scholar] [CrossRef]
- Zienkiewicz, M.; Krupnik, T.; Drożak, A.; Kania, K. PEG-mediated, Stable, Nuclear and Chloroplast Transformation of Cyanidioschizon merolae. Bio-Protocol 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Jin, E.S.; Polle, J.E.W.; Melis, A. Involvement of zeaxanthin and of the Cbr protein in the repair of photosystem II from photoinhibition in the green alga Dunaliella salina. Biochim. Biophys. Acta-Bioenerg. 2001, 1506, 244–259. [Google Scholar] [CrossRef] [Green Version]
- Economou, C.; Wannathong, T.; Szaub, J.; Purton, S. A simple, low-cost method for chloroplast transformation of the green alga Chlamydomonas reinhardtii. Methods Mol. Biol. 2014, 1132, 401–411. [Google Scholar] [CrossRef]
- Kindle, K.L.; Richards, K.L.; Stern, D.B. Engineering the chloroplast genome: Techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1991, 88, 1721–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchanan, M.J.; Snell, W.J. Biochemical studies on lysin, a cell wall degrading enzyme released during fertilization in Chlamydomonas. Exp. Cell Res. 1988. [Google Scholar] [CrossRef]
- Zorin, B.; Hegemann, P.; Sizova, I. Nuclear-gene targeting by using single-stranded DNA avoids illegitimate DNA integration in Chlamydomonas reinhardtii. Eukaryot. Cell 2005, 4, 1264–1272. [Google Scholar] [CrossRef] [Green Version]
- Rivera, A.L.; Magaña-Ortíz, D.; Gómez-Lim, M.; Fernández, F.; Loske, A.M. Physical methods for genetic transformation of fungi and yeast. Phys. Life Rev. 2014, 11, 184–203. [Google Scholar] [CrossRef]
- Dunahay, T.G. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. Biotechniques 1993, 15, 452–460. [Google Scholar] [PubMed]
- Hiramatsu, S.; Ishihara, M.; Fujie, M.; Usami, S. Expression of a chitinase gene and lysis of the host cell wall during Chlorella virus CVK2 infection. Virology 1999, 260, 308–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Gurnon, J.R.; Adams, B.J.; Graves, M.V.; Van Etten, J.L. Characterization of a β-1,3-glucanase encoded by Chlorella virus PBCV-1. Virology 2000, 276, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Sugimoto, I.; Hiramatsu, S.; Murakami, D.; Fujie, M.; Usami, S.; Yamada, T. Algal-lytic activities encoded by Chlorella virus CVK2. Virology 2000, 277, 119–126. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol. 2002, 4, 63–73. [Google Scholar] [CrossRef]
- Maruyama, M.; Horáková, I.; Honda, H.; Xing, X.H.; Shiragami, N.; Unno, H. Introduction of foreign DNA into Chlorella saccharophila by electroporation. Biotechnol. Tech. 1994, 8, 821–826. [Google Scholar] [CrossRef]
- Jarvis, E.E.; Brown, L.M. Transient expression of firefly luciferase in protoplasts of the green alga Chlorella ellipsoidea. Curr. Genet. 1991, 19, 317–321. [Google Scholar] [CrossRef]
- Shimogawara, K.; Fujiwara, S.; Grossman, A.; Usuda, H. High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 1998, 148, 1821–1828. [Google Scholar] [CrossRef]
- Brown, L.E.; Sprecher, S.L.; Keller, L.R. Introduction of exogenous DNA into Chlamydomonas reinhardtii by electroporation. Mol. Cell. Biol. 1991, 11, 2328–2332. [Google Scholar] [CrossRef] [Green Version]
- Weaver, J.C. Electroporation theory. Concepts and mechanisms. Methods Mol. Biol. 1995, 48, 3–28. [Google Scholar] [PubMed]
- Ladygin, V.G. Efficient transformation of mutant cells of Chlamydomonas reinhardtii by electroporation. Process Biochem. 2004, 39, 1685–1691. [Google Scholar] [CrossRef]
- Azencott, H.R.; Peter, G.F.; Prausnitz, M.R. Influence of the Cell Wall on Intracellular Delivery to Algal Cells by Electroporation and Sonication. Ultrasound Med. Biol. 2007, 33, 1805–1817. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.K.H.; Qiao, S.Y.; Wu, M. Insertion mutagenesis of Chlamydomonas reinhardtii by electroporation and heterologous DNA. Biochem. Mol. Biol. Int. 1995, 36, 1025–1035. [Google Scholar] [PubMed]
- Chen, Y.; Hu, H. High efficiency transformation by electroporation of the freshwater alga Nannochloropsis limnetica. World J. Microbiol. Biotechnol. 2019, 35, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hu, H. High-efficiency nuclear transformation of the diatom Phaeodactylum tricornutum by electroporation. Mar. Genom. 2014, 16, 63–66. [Google Scholar] [CrossRef]
- Holmqvist, M.; Stensjö, K.; Oliveira, P.; Lindberg, P.; Lindblad, P. Characterization of the hupSL promoter activity in Nostoc punctiforme ATCC 29133. BMC Microbiol. 2009, 9. [Google Scholar] [CrossRef] [Green Version]
- Thiel, T.; Poo, H. Transformation of a filamentous cyanobacterium by electroporation. J. Bacteriol. 1989, 171, 5743–5746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaeger, D.; Hübner, W.; Huser, T.; Mussgnug, J.H.; Kruse, O. Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum. J. Biotechnol. 2017, 249, 10–15. [Google Scholar] [CrossRef]
- Doron, L.; Segal, N.; Shapira, M. Transgene expression in microalgae—from tools to applications. Front. Plant Sci. 2016, 7, 505. [Google Scholar] [CrossRef] [PubMed]
- Yamano, T.; Iguchi, H.; Fukuzawa, H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 2013, 115, 691–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Yang, L.; Wen, X.; Chen, Z.; Liang, Q.; Li, J.; Wang, W. Rapid and high efficiency transformation of Chlamydomonas reinhardtii by square-wave electroporation. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Angstenberger, M.; De Signori, F.; Vecchi, V.; Dall’Osto, L.; Bassi, R. Cell Synchronization Enhances Nuclear Transformation and Genome Editing via Cas9 Enabling Homologous Recombination in Chlamydomonas reinhardtii. ACS Synth. Biol. 2020, 9, 2840–2850. [Google Scholar] [CrossRef] [PubMed]
- Im, D.J.; Jeong, S.-N.; Yoo, B.S.; Kim, B.; Kim, D.-P.; Jeong, W.-J.; Kang, I.S. Digital Microfluidic Approach for Efficient Electroporation with High Productivity: Transgene Expression of Microalgae without Cell Wall Removal. Anal. Chem. 2015, 87, 6592–6599. [Google Scholar] [CrossRef]
- Bodénès, P.; Wang, H.-Y.; Lee, T.-H.; Chen, H.-Y.; Wang, C.-Y. Microfluidic techniques for enhancing biofuel and biorefinery industry based on microalgae. Biotechnol. Biofuels 2019, 12, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boynton, J.E.; Gillham, N.W.; Harris, E.H.; Hosler, J.P.; Johnson, A.M.; Jones, A.R.; Randolph-Anderson, B.L.; Robertson, D.; Klein, T.M.; Shark, K.B.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef]
- Day, A.; Debuchy, R.; van Dillewijn, J.; Purton, S.; Rochaix, J.-D. Studies on the maintenance and expression of cloned DNA fragments in the nuclear genome of the green alga Chlamydomonas reinhardtii. Physiol. Plant. 1990, 78, 254–260. [Google Scholar] [CrossRef]
- Remacle, C.; Cardol, P.; Coosemans, N.; Gaisne, M.; Bonnefoy, N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl. Acad. Sci. USA 2006, 103, 4771–4776. [Google Scholar] [CrossRef] [Green Version]
- Altpeter, F.; Baisakh, N.; Beachy, R.; Bock, R.; Capell, T.; Christou, P.; Daniell, H.; Datta, K.; Datta, S.; Dix, P.J.; et al. Particle bombardment and the genetic enhancement of crops: Myths and realities. Mol. Breed. 2005, 15, 305–327. [Google Scholar] [CrossRef]
- Li, Z.; Bock, R. Replication of bacterial plasmids in the nucleus of the red alga Porphyridium purpureum. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Matamoros, M.F.; Villanueva, M.A.; Islas-Flores, T. Genetic transformation of cell-walled plant and algae cells: Delivering DNA through the cell wall. Brief. Funct. Genom. 2018, 17, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Schiedlmeier, B.; Schmitt, R.; Müller, W.; Kirk, M.M.; Gruber, H.; Mages, W.; Kirk, D.L. Nuclear transformation of Volvox carteri. Proc. Natl. Acad. Sci. USA 1994, 91, 5080–5084. [Google Scholar] [CrossRef] [Green Version]
- Feng, S.; Xue, L.; Liu, H.; Lu, P. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol. Biol. Rep. 2009, 36, 1433–1439. [Google Scholar] [CrossRef] [PubMed]
- Apt, K.E.; Grossman, A.R.; Kroth-Pancic, P.G. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol. Gen. Genet. MGG 1996, 252, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Randolph-Anderson, B.; Boynton, J.E.; Dawson, J.; Dunder, E.; Eskes, R.; Gillham, N.W.; Johnson, A.; Perlman, P.S.; Suttie, J.; Heiser, W.C. Sub-Micron Gold Particles are Superior to Larger Particles for Efficient Biolistic Transformation of Organelles and Some Cell Types. 1995. Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_2015.pdf (accessed on 31 January 2021).
- Martin-Ortigosa, S.; Wang, K. Proteolistics: A biolistic method for intracellular delivery of proteins. Transgenic Res. 2014, 23, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Serif, M.; Dubois, G.; Finoux, A.L.; Teste, M.A.; Jallet, D.; Daboussi, F. One-step generation of multiple gene knock-outs in the diatom Phaeodactylum tricornutum by DNA-free genome editing. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayfield, S.P.; Kindle, K.L. Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc. Natl. Acad. Sci. USA 1990, 87, 2087–2091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramesh, V.M.; Bingham, S.E.; Webber, A.N. A simple method for chloroplast transformation in Chlamydomonas reinhardtii. Methods Mol. Biol. 2011, 684, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Stevens, S.E.; Porter, R.D. Heterospecific transformation among cyanobacteria. J. Bacteriol. 1986, 167, 1074–1076. [Google Scholar] [CrossRef] [Green Version]
- Stucken, K.; Koch, R.; Dagan, T. Cyanobacterial defense mechanisms against foreign DNA transfer and their impact on genetic engineering. Biol. Res. 2013, 46, 373–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shestakov, S.V.; Khyen, N.T. Evidence for genetic transformation in blue-green alga Anacystis nidulans. MGG Mol. Gen. Genet. 1970, 107, 372–375. [Google Scholar] [CrossRef]
- Grigorieva, G.; Shestakov, S. Transformation in the cyanobacterium Synechocystis sp. 6803. FEMS Microbiol. Lett. 1982, 13, 367–370. [Google Scholar] [CrossRef]
- Onai, K.; Morishita, M.; Kaneko, T.; Tabata, S.; Ishiura, M. Natural transformation of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1: A simple and efficient method for gene transfer. Mol. Genet. Genom. 2004, 271, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef]
- Williams, J.G.K.; Szalay, A.A. Stable integration of foreign DNA into the chromosome of the cyanobacterium Synechococcus R2. Gene 1983, 24, 37–51. [Google Scholar] [CrossRef]
- Yoshihara, S.; Geng, X.X.; Okamoto, S.; Yura, K.; Murata, T.; Go, M.; Ohmori, M.; Ikeuchi, M. Mutational analysis of genes involved in pilus structure, motility and transformation competency in the unicellular motile cyanobacterium Synechocystis sp. PCC 6803. Plant Cell Physiol. 2001, 42, 63–73. [Google Scholar] [CrossRef] [Green Version]
- Nakasugi, K.; Svenson, C.J.; Neilan, B.A. The competence gene, comF, from Synechocystis sp. strain PCC 6803 is involved in natural transformation, phototactic motility and piliation. Microbiology 2006, 152, 3623–3631. [Google Scholar] [CrossRef] [Green Version]
- Wendt, K.E.; Pakrasi, H.B. Genomics approaches to deciphering natural transformation in cyanobacteria. Front. Microbiol. 2019, 10, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Proels, R. Stable Transformation of Cyanobacterium Synechocystis sp. Bio-Protocol 2014, 4. [Google Scholar] [CrossRef]
- Nagarajan, A.; Winter, R.; Eaton-Rye, J.; Burnap, R. A synthetic DNA and fusion PCR approach to the ectopic expression of high levels of the D1 protein of photosystem II in Synechocystis sp. PCC 6803. J. Photochem. Photobiol. B Biol. 2011, 104, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.V.; Martens, S.B.B.; Lanes, C.F.C.; Marins, L.F. Improved genetic transformation of Synechococcus elongatus PCC 7942 using linear DNA fragments in association with a DNase inhibitor. Biotechnol. Res. Innov. 2017, 1, 123–128. [Google Scholar] [CrossRef]
- Vioque, A. Transformation of cyanobacteria. Adv. Exp. Med. Biol. 2007, 616, 12–22. [Google Scholar] [PubMed]
- Karas, B.J.; Diner, R.E.; Lefebvre, S.C.; McQuaid, J.; Phillips, A.P.R.; Noddings, C.M.; Brunson, J.K.; Valas, R.E.; Deerinck, T.J.; Jablanovic, J.; et al. Designer diatom episomes delivered by bacterial conjugation. Nat. Commun. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Currier, T.C.; Wolk, C.P. Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J. Bacteriol. 1979. [Google Scholar] [CrossRef] [Green Version]
- Thiel, T.; Peter Wolk, C. Conjugal Transfer of Plasmids to Cyanobacteria. Methods Enzymol. 1987, 153, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Wolk, C.P.; Vonshak, A.; Kehoe, P.; Elhai, J. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Isotopenpraxis 1984, 20, 1561–1565. [Google Scholar] [CrossRef] [Green Version]
- Brahamsha, B. A Genetic Manipulation System for Oceanic Cyanobacteria of the Genus Synechococcus. Appl. Environ. Microbiol. 1996, 62, 1747–1751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tolonen, A.C.; Liszt, G.B.; Hess, W.R. Genetic Manipulation of Prochlorococcus Strain MIT9313: Green Fluorescent Protein Expression from an RSF1010 Plasmid and Tn5 Transposition. Appl. Environ. Microbiol. 2006, 72, 7607–7613. [Google Scholar] [CrossRef] [Green Version]
- Marraccini, P.; Bulteau, S.; Cassier-Chauvat, C.; Mermet-Bouvier, P.; Chauvat, F. A conjugative plasmid vector for promoter analysis in several cyanobacteria of the genera Synechococcus and Synechocystis. Plant Mol. Biol. 1993, 23, 905–909. [Google Scholar] [CrossRef]
- Tsinoremas, N.F.; Kutach, A.K.; Strayer, C.A.; Golden, S.S. Efficient gene transfer in Synechococcus sp. strains PCC 7942 and PCC 6301 by interspecies conjugation and chromosomal recombination. J. Bacteriol. 1994, 176, 6764–6768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, C.F.; Sturme, M.H.J.; D’Adamo, S.; Weusthuis, R.A.; Wijffels, R.H. Stable transformation of the green algae Acutodesmus obliquus and Neochloris oleoabundans based on E. coli conjugation. Algal Res. 2019, 39, 101453. [Google Scholar] [CrossRef]
- Fabris, M.; George, J.; Kuzhiumparambil, U.; Lawson, C.A.; Jaramillo-Madrid, A.C.; Abbriano, R.M.; Vickers, C.E.; Ralph, P. Extrachromosomal Genetic Engineering of the Marine Diatom Phaeodactylum tricornutum Enables the Heterologous Production of Monoterpenoids. ACS Synth. Biol. 2020. [Google Scholar] [CrossRef]
- Dunahay, T.G.; Jarvis, E.E.; Roessler, P.G. Genetic Transformation of the Diatoms Cyclotella Cryptica and Navicula Saprophila. J. Phycol. 1995, 31, 1004–1012. [Google Scholar] [CrossRef]
- Falciatore, A.; Casotti, R.; Leblanc, C.; Abrescia, C.; Bowler, C. Transformation of Nonselectable Reporter Genes in Marine Diatoms. Mar. Biotechnol. 1999, 1, 239–251. [Google Scholar] [CrossRef]
- Miyagawa, A.; Okami, T.; Kira, N.; Yamaguchi, H.; Ohnishi, K.; Adachi, M. Research note: High efficiency transformation of the diatom Phaeodactylum tricornutum with a promoter from the diatom Cylindrotheca fusiformis. Phycol. Res. 2009, 57, 142–146. [Google Scholar] [CrossRef]
- Diner, R.E.; Bielinski, V.A.; Dupont, C.L.; Allen, A.E.; Weyman, P.D. Refinement of the diatom episome maintenance sequence and improvement of conjugation-based DNA delivery methods. Front. Bioeng. Biotechnol. 2016, 4, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, M.F.; Wallis, J.G.; Campbell, E.L.; Meeks, J.C. Transposon mutagenesis of Nostoc sp. strain ATCC 29133, a filamentous cyanobacterium with multiple cellular differentiation alternatives. Microbiology 1994, 140, 3233–3240. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.K.; Nymark, M.; Sparstad, T.; Bones, A.M.; Winge, P. Transgene-free genome editing in marine algae by bacterial conjugation—comparison with biolistic CRISPR/Cas9 transformation. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Poliner, E.; Clark, E.; Cummings, C.; Benning, C.; Farre, E.M. A high-capacity gene stacking toolkit for the oleaginous microalga, Nannochloropsis oceanica CCMP1779. Algal Res. 2020, 45, 101664. [Google Scholar] [CrossRef]
- Poliner, E.; Takeuchi, T.; Du, Z.Y.; Benning, C.; Farre, E.M. Non-transgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779 Eric. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Tzfira, T.; Citovsky, V. Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr. Opin. Biotechnol. 2006, 17, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Kunik, T. Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. USA 2001, 98, 1871–1876. [Google Scholar] [CrossRef] [PubMed]
- Schell, J.; Van Montagu, M. The Ti-plasmid of Agrobacterium tumefaciens, a natural vector for the introduction of nif genes in plants? Basic Life Sci. 1977, 9, 159–179. [Google Scholar] [PubMed]
- Zambryski, P.; Joos, H.; Genetello, C.; Leemans, J.; Van Montagu, M.; Schell, J. Ti plasmid vector for the introduction of DNA into plant cells without alteration of their normal regeneration capacity. EMBO J. 1983, 2, 2143–2150. [Google Scholar] [CrossRef]
- Hamilton, C.M.; Frary, A.; Lewis, C.; Tanksley, S.D. Stable transfer of intact high molecular weight DNA into plant chromosomes. Proc. Natl. Acad. Sci. USA 1996, 93, 9975–9979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valvekens, D.; Montagu, M.V.; Lijsebettens, M.V. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc. Natl. Acad. Sci. USA 1988, 85, 5536–5540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.V.; Misquitta, R.W.; Reddy, V.S.; Rao, B.J.; Rajam, M.V. Genetic transformation of the green alga - Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci. 2004, 166, 731–738. [Google Scholar] [CrossRef]
- Kathiresan, S.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (chlorophyceae, volvocales). J. Phycol. 2009, 45, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.S.; Yee, W.; Aziz, A. Assessment of factors affecting Agrobacterium-mediated genetic transformation of the unicellular green alga, Chlorella vulgaris. World J. Microbiol. Biotechnol. 2012, 28, 1771–1779. [Google Scholar] [CrossRef]
- Rathod, J.P.; Prakash, G.; Pandit, R.; Lali, A.M. Agrobacterium-mediated transformation of promising oil-bearing marine algae Parachlorella kessleri. Photosynth. Res. 2013, 118, 141–146. [Google Scholar] [CrossRef]
- Anila, N.; Chandrashekar, A.; Ravishankar, G.A.; Sarada, R. Establishment of Agrobacterium tumefaciens -mediated genetic transformation in Dunaliella bardawil. Eur. J. Phycol. 2011, 46, 36–44. [Google Scholar] [CrossRef] [Green Version]
- Pratheesh, P.T.; Vineetha, M.; Kurup, G.M. An efficient protocol for the Agrobacterium-mediated genetic transformation of microalga Chlamydomonas reinhardtii. Mol. Biotechnol. 2014, 56, 507–515. [Google Scholar] [CrossRef]
- Mini, P.; Demurtas, O.C.; Valentini, S.; Pallara, P.; Aprea, G.; Ferrante, P.; Giuliano, G. Agrobacterium-mediated and electroporation-mediated transformation of Chlamydomonas reinhardtii: A comparative study. BMC Biotechnol. 2018, 18, 11. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.; Lein, W.; Thiyam, G.; Lindenberger, C.P.; Buchholz, R.; Vadakedath, N. Stable nuclear transformation of rhodophyte species Porphyridium purpureum: Advanced molecular tools and an optimized method. Photosynth. Res. 2019, 140, 173–188. [Google Scholar] [CrossRef]
- Michielse, C.B.; Hooykaas, P.J.J.; van den Hondel, C.A.M.J.J.; Ram, A.F.J. Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 2005, 48, 1–17. [Google Scholar] [CrossRef]
- Lauersen, K.J. Eukaryotic microalgae as hosts for light-driven heterologous isoprenoid production. Planta 2019, 249, 155–180. [Google Scholar] [CrossRef]
- Zhang, R.; Patena, W.; Armbruster, U.; Gang, S.S.; Blum, S.R.; Jonikas, M.C. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 2014, 26, 1398–1409. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Cliften, P.F.; Dutcher, S.K. MAPINS, a highly efficient detection method that identifies insertional mutations and complex DNA rearrangements. Plant Physiol. 2018, 178, 1436–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rádis-Baptista, G.; Campelo, I.S.; Morlighem, J.É.R.L.; Melo, L.M.; Freitas, V.J.F. Cell-penetrating peptides (CPPs): From delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J. Biotechnol. 2017, 252, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Lehto, T.; Ezzat, K.; Wood, M.J.A.; EL Andaloussi, S. Peptides for nucleic acid delivery. Adv. Drug Deliv. Rev. 2016, 106, 172–182. [Google Scholar] [CrossRef]
- Margus, H.; Padari, K.; Pooga, M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol. Ther. 2012, 20, 525–533. [Google Scholar] [CrossRef] [Green Version]
- Mishra, V.K.; Anantharamaiah, G.M.; Segrest, J.P.; Palgunachari, M.N.; Chaddha, M.; Sham, S.W.S.; Krishna, N.R. Association of a Model Class A (Apolipoprotein) Amphipathic α Helical Peptide with Lipid. J. Biol. Chem. 2006, 281, 6511–6519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Carneado, J.; Kogan, M.J.; Pujals, S.; Giralt, E. Amphipathic Peptides and Drug Delivery. Proc. Biopolym. Pept. Sci. Sect. 2004, 76, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Morris, M.C.; Vidal, P.; Chaloin, L.; Heitz, F.; Divita, G. A new peptide vector for efficient delivery of oligonucleotides into mammalian cells. Nucleic Acids Res. 1997, 25, 2730–2736. [Google Scholar] [CrossRef] [PubMed]
- Chugh, A.; Eudes, F. Cellular uptake of cell-penetrating peptides pVEC and transportan in plants. J. Pept. Sci. 2008, 14, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Oehlke, J.; Scheller, A.; Wiesner, B.; Krause, E.; Beyermann, M.; Klauschenz, E.; Melzig, M.; Bienert, M. Cellular uptake of an α-helical amphipathic model peptide with the potential to deliver polar compounds into the cell interior non-endocytically. Biochim. Biophys. Acta-Biomembr. 1998. [Google Scholar] [CrossRef] [Green Version]
- Gräslund, A.; Madani, F.; Lindberg, S.; Langel, Ü.; Futaki, S. Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011, 2011, 414729. [Google Scholar]
- Pooga, M.; Langel, Ü. Classes of cell-penetrating peptides. In Cell-Penetrating Peptides: Methods and Protocols; Springer: New York, NY, USA, 2015; pp. 3–28. ISBN 9781493928064. [Google Scholar]
- Fuselier, T.; Wimley, W.C. Spontaneous Membrane Translocating Peptides: The Role of Leucine-Arginine Consensus Motifs. Biophys. J. 2017, 113, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Vivès, E.; Brodin, P.; Lebleu, B. A truncated HIV-1 Tat protein basic domain rapidly translocates through the plasma membrane and accumulates in the cell nucleus. J. Biol. Chem. 1997, 272, 16010–16017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Derossi, D.; Joliot, A.H.; Chassaing, G.; Prochiantz, A. The third helix of the Antennapedia homeodomain translocates through biological membranes. J. Biol. Chem. 1994, 269, 10444–10450. [Google Scholar] [CrossRef]
- Rojas, M.; Donahue, J.P.; Tan, Z.; Lin, Y.Z. Genetic engineering of proteins with cell membrane permeability. Nat. Biotechnol. 1998, 16, 370–375. [Google Scholar] [CrossRef]
- Liu, B.R.; Huang, Y.W.; Lee, H.J. Mechanistic studies of intracellular delivery of proteins by cell-penetrating peptides in cyanobacteria. BMC Microbiol. 2013, 13, 57. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Niu, J.; Huan, L.; Huang, A.; He, L.; Wang, G. Cell penetrating peptide can transport dsRNA into microalgae with thin cell walls. Algal Res. 2015, 8, 135–139. [Google Scholar] [CrossRef]
- Frankel, A.D.; Pabo, C.O. Cellular uptake of the tat protein from human immunodeficiency virus. Cell 1988, 55, 1189–1193. [Google Scholar] [CrossRef]
- Hyman, J.M.; Geihe, E.I.; Trantow, B.M.; Parvin, B.; Wender, P.A. A molecular method for the delivery of small molecules and proteins across the cell wall of algae using molecular transporters. Proc. Natl. Acad. Sci. USA 2012, 109, 13225–13230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadamchetty, P.; Mullapudi, P.L.V.; Sanagala, R.; Markandan, M.; Polumetla, A.K. Genetic transformation of Chlorella vulgaris mediated by HIV-TAT peptide. 3 Biotech 2019, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Suresh, A.; Kim, Y.C. Translocation of cell penetrating peptides on Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2013, 110, 2795–2801. [Google Scholar] [CrossRef]
- Holm, T.; Netzereab, S.; Hansen, M.; Langel, Ü.; Hällbrink, M. Uptake of cell-penetrating peptides in yeasts. FEBS Lett. 2005, 579, 5217–5222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joliot, A.; Pernelle, C.; Deagostini-Bazin, H.; Prochiantz, A. Antennapedia homeobox peptide regulates neural morphogenesis. Proc. Natl. Acad. Sci. USA 1991, 88, 1864–1868. [Google Scholar] [CrossRef] [Green Version]
- Kang, S.; Jeon, S.; Kim, S.; Chang, Y.K.; Kim, Y.C. Development of a pVEC peptide-based ribonucleoprotein (RNP) delivery system for genome editing using CRISPR/Cas9 in Chlamydomonas reinhardtii. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Suresh, A.; Kim, Y.C. A highly efficient cell penetrating peptide pVEC-mediated protein delivery system into microalgae. Algal Res. 2017, 24, 360–367. [Google Scholar] [CrossRef]
- Boussif, O.; LezoualC’H, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [Green Version]
- Haensler, J.; Szoka, F.C. Polyamidoamine Cascade Polymers Mediate Efficient Transfection of Cells in Culture. Bioconjug. Chem. 1993, 4, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Benjaminsen, R.V.; Mattebjerg, M.A.; Henriksen, J.R.; Moghimi, S.M.; Andresen, T.L. The possible “proton sponge” effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol. Ther. 2013, 21, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Pack, D.W.; Hoffman, A.S.; Pun, S.; Stayton, P.S. Design and development of polymers for gene delivery. Nat. Rev. Drug Discov. 2005, 4, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Klibanov, A.M. Enhancing polyethylenimine’s delivery of plasmid DNA into mammalian cells. Proc. Natl. Acad. Sci. USA 2002, 99, 14640–14645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, M.L.; Meister, G.E.; Koerber, J.T.; Pack, D.W. Partial Acetylation of Polyethylenimine Enhances In Vitro Gene Delivery. Pharm. Res. 2004, 21, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Carboni, V.; Maaliki, C.; Alyami, M.; Alsaiari, S.; Khashab, N. Synthetic Vehicles for Encapsulation and Delivery of CRISPR/Cas9 Gene Editing Machinery. Adv. Ther. 2019, 2, 1800085. [Google Scholar] [CrossRef]
- Furukawa, H.; Cordova, K.E.; O’Keeffe, M.; Yaghi, O.M. The chemistry and applications of metal-organic frameworks. Science 2013, 341. [Google Scholar] [CrossRef] [Green Version]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef] [Green Version]
- Alyami, M.Z.; Alsaiari, S.K.; Li, Y.; Qutub, S.S.; Aleisa, F.A.; Sougrat, R.; Merzaban, J.S.; Khashab, N.M. Cell-Type-Specific CRISPR/Cas9 Delivery by Biomimetic Metal Organic Frameworks. J. Am. Chem. Soc. 2020, 142, 1715–1720. [Google Scholar] [CrossRef]
- Patil, A.J.; Muthusamy, E.; Mann, S. Synthesis and Self-Assembly of Organoclay-Wrapped Biomolecules. Angew. Chemie Int. Ed. 2004, 43, 4928–4933. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.J.; Li, M.; Dujardin, E.; Mann, S. Novel bioinorganic nanostructures based on mesolamellar intercalation or single-molecule wrapping of DNA using organoclay building blocks. Nano Lett. 2007, 7, 2660–2665. [Google Scholar] [CrossRef] [PubMed]
- Mann, S. Self-assembly and transformation of hybrid nano-objects and nanostructures under equilibrium and non-equilibrium conditions. Nat. Mater. 2009, 8, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, Y.-C.; Cho, D.-H.; Lee, H.U.; Huh, Y.S.; Kim, G.-J.; Kim, H.-S. A Simple and Non-Invasive Method for Nuclear Transformation of Intact-walled Chlamydomonas reinhardtii. PLoS ONE 2014, 9, e101018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, S.; Burkett, S.L.; Davis, S.A.; Fowler, C.E.; Mendelson, N.H.; Sims, S.D.; Walsh, D.; Whilton, N.T. Sol-Gel Synthesis of Organized Matter. Chem. Mater. 1997, 9, 2300–2310. [Google Scholar] [CrossRef]
- Han, H.K.; Lee, Y.C.; Lee, M.Y.; Patil, A.J.; Shin, H.J. Magnesium and calcium organophyllosilicates: Synthesis and in vitro cytotoxicity study. ACS Appl. Mater. Interfaces 2011, 3, 2564–2572. [Google Scholar] [CrossRef]
- Kim, J.; Grate, J.W. Single-Enzyme Nanoparticles Armored by a Nanometer-Scale Organic/Inorganic Network. Nano Lett. 2003, 3, 1219–1222. [Google Scholar] [CrossRef]
- Ichinose, I.; Hashimoto, Y.; Kunitake, T. Wrapping of Bio-macromolecules (Dextran, Amylopectin, and Horse Heart Cytochrome c) with Ultrathin Silicate Layer. Chem. Lett. 2004, 33, 656–657. [Google Scholar] [CrossRef]
- Numata, M.; Li, C.; Bae, A.H.; Kaneko, K.; Sakurai, K.; Shinkai, S. Β-1,3-Glucan Polysaccharide Can Act As a One-Dimensional Host To Create Novel Silica Nanofiber Structures. Chem. Commun. 2005, 4655–4657. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Eddaoudi, M.; O’Keeffe, M.; Yaghi, O.M. Design and synthesis of an exceptionally stable and highly porous metal- organic framework. Nature 1999, 402, 276–279. [Google Scholar] [CrossRef] [Green Version]
- Phan, A.; Doonan, C.J.; Uribe-Romo, F.J.; Knobler, C.B.; Okeeffe, M.; Yaghi, O.M. Synthesis, structure, and carbon dioxide capture properties of zeolitic imidazolate frameworks. Acc. Chem. Res. 2010, 43, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Venna, S.R.; Jasinski, J.B.; Carreon, M.A. Structural evolution of zeolitic imidazolate framework-8. J. Am. Chem. Soc. 2010, 132, 18030–18033. [Google Scholar] [CrossRef]
- Liang, K.; Ricco, R.; Doherty, C.M.; Styles, M.J.; Bell, S.; Kirby, N.; Mudie, S.; Haylock, D.; Hill, A.J.; Doonan, C.J.; et al. Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Shieh, F.K.; Wang, S.C.; Yen, C.I.; Wu, C.C.; Dutta, S.; Chou, L.Y.; Morabito, J.V.; Hu, P.; Hsu, M.H.; Wu, K.C.W.; et al. Imparting Functionality to Biocatalysts via Embedding Enzymes into Nanoporous Materials by a de Novo Approach: Size-Selective Sheltering of Catalase in Metal-Organic Framework Microcrystals. J. Am. Chem. Soc. 2015, 137, 4276–4279. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.L.; Newman, C.; Briggs, S.; Seymour, L.; Sheridan, P.J. Nonviral gene delivery: Techniques and implications for molecular medicine. Expert Rev. Mol. Med. 2003, 5. [Google Scholar] [CrossRef]
- Balazs, D.A.; Godbey, W.T. Liposomes for Use in Gene Delivery. J. Drug Deliv. 2011, 2011, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hug, P.; Sleight, R.G. Liposomes for the transformation of eukaryotic cells. BBA-Mol. Basis Dis. 1991, 1097, 1–17. [Google Scholar] [CrossRef]
- Gad, A.E.; Rosenberg, N.; Altman, A. Liposome-mediated gene delivery into plant cells. Physiol. Plant. 1990, 79, 177–183. [Google Scholar] [CrossRef]
- Thierry, A.R.; Rabinovich, P.; Peng, B.; Mahan, L.C.; Bryant, J.L.; Gallo, R.C. Characterization of liposome-mediated gene delivery: Expression, stability and pharmacokinetics of plasmid DNA. Gene Ther. 1997, 4, 226–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 8–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jesorka, A.; Orwar, O. Liposomes: Technologies and Analytical Applications. Annu. Rev. Anal. Chem. 2008, 1, 801–832. [Google Scholar] [CrossRef]
- Ohta, K.; Ichihashi, N. Liposome fragment-mediated introduction of multiple plasmids into Bacillus subtilis. Biochem. Biophys. Reports 2019, 18, 100646. [Google Scholar] [CrossRef] [PubMed]
- Yachi, K.; Harashima, H.; Kikuchi, H.; Sudo, R.; Yamauchi, H.; Ebihara, K.; Matsuo, H.; Funato, K.; Kiwada, H. Biopharmaceutical evaluation of the liposomes prepared by rehydration of freeze-dried empty liposomes (FDELs) with an aqueous solution of a drug. Biopharm. Drug Dispos. 1996, 17, 589–605. [Google Scholar] [CrossRef]
- Mannino, R.J.; Gould-Fogerite, S. Liposome mediated gene transfer. Biotechniques 1988, 6, 682–690. [Google Scholar] [CrossRef]

| Method | Species | Advantage | Disadvantage | Transformation Efficiency (cells/µg DNA) | Initial Cell Concentration (cells/mL) | DNA Added (µg) | Ref. |
|---|---|---|---|---|---|---|---|
| Glass bead agitation and PEG mediated DNA delivery | C. reinhardtii | Simple; inexpensive; fast | Requires cell wall removal/deficiency; occasional genome lesions | 103 | 108 | 2 | [14,15] |
| C. merolae | |||||||
| C. vulgaris | |||||||
| C. ellipsoidea | |||||||
| D. salina | |||||||
| protoplasts | |||||||
| Electroporation | C. reinhardtii | Not affected by cell wall presence; Occasional genome lesions | Specialized equipment | 105 | 108 | 2.5 | [34,37,46,48] |
| M. neglectum | |||||||
| Nannochloropsis sp. | |||||||
| P. tricornutum | |||||||
| Anabaena sp. | |||||||
| N. punctiforme | |||||||
| N. limnetica | |||||||
| Digital microfluidic electroporation (DME) | C. reinhardtii | Not affected by cell wall presence; occasional genome lesions | Specialized equipment | 104 | 106 | 1 | [49] |
| Square electric pulse electroporation | C. reinhardtii | Not affected by cell wall presence; occasional genome lesions | Specialized equipment | 103 | 107 | 0.1 | [47] |
| Microparticle bombardment (gene gun) | C. reinhardtii | Plastid target; not affected by cell wall | Cell viability compromise; specialized equipment | 102 | 105 | 0.1 | [23,51,52,53,59] |
| P. purpureum | |||||||
| D. salina | |||||||
| V. carteri | |||||||
| P. tricornutum | |||||||
| Natural transformation | Anacystis nidulans | Straightforward method for extensive genetic engineering | Limited to some species | 104 | 107 | 5 | [67,68,75] |
| Synechocystis sp. | |||||||
| Synechococcus sp. | |||||||
| T. elongatus | |||||||
| Bacterial conjugation | Anabaena | Low non-target insertions/knockouts; independent episome replication; allows delivery of large DNA fragments | Relies on target species characteristics based on recipient capability to integrate or maintain the vector | 104–106 | 107–109 | 30–50 | [81,82,84,85,86] |
| Nostoc sp. | |||||||
| Prochlorococcus sp. | |||||||
| Synechococcus sp. | |||||||
| Synechocystis sp. | |||||||
| N. punctiforme | |||||||
| P. tricornutum | |||||||
| T. pseudonana | |||||||
| A. obliquus | |||||||
| N. oleoabundans | |||||||
| N. oceanica | |||||||
| Agrobacterium-mediated transformation | C. reinhardtii | Low gene rearrangements; low foreign transcript silencing | Labor-intensive; no higher gene expression reported | 10 | 108 | 30 | [103,109] |
| H. lacustris | |||||||
| Chlorella sp. | |||||||
| D. bardawil | |||||||
| Symbiodinium sp. | |||||||
| Nannochloropsis sp. | |||||||
| P. kessleri | |||||||
| Cell-Penetrating Peptides | Synechocystis sp. | High cargo stability; internalized efficiently | Requires cell wall removal/deficiency; optimized for mammalian cells | 104 | 105–106 | 10–50 | [129,130,132] |
| S. elongatus | |||||||
| C. reinhardtii | |||||||
| C. vulgaris | |||||||
| P. tricornutum | |||||||
| D. salina | |||||||
| N. oleoabundans | |||||||
| S. dimorphus | |||||||
| Botrycoccus braunii | |||||||
| Metal-Organic Frameworks (MOF) | C. reinhardtii | High aqueous stability pH-buffering capacity, versatile | Not yet optimized requires cell wall removal/deficiency | 102 | 106 | 0.7 | [152] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gutiérrez, S.; Lauersen, K.J. Gene Delivery Technologies with Applications in Microalgal Genetic Engineering. Biology 2021, 10, 265. https://doi.org/10.3390/biology10040265
Gutiérrez S, Lauersen KJ. Gene Delivery Technologies with Applications in Microalgal Genetic Engineering. Biology. 2021; 10(4):265. https://doi.org/10.3390/biology10040265
Chicago/Turabian StyleGutiérrez, Sergio, and Kyle J. Lauersen. 2021. "Gene Delivery Technologies with Applications in Microalgal Genetic Engineering" Biology 10, no. 4: 265. https://doi.org/10.3390/biology10040265
APA StyleGutiérrez, S., & Lauersen, K. J. (2021). Gene Delivery Technologies with Applications in Microalgal Genetic Engineering. Biology, 10(4), 265. https://doi.org/10.3390/biology10040265