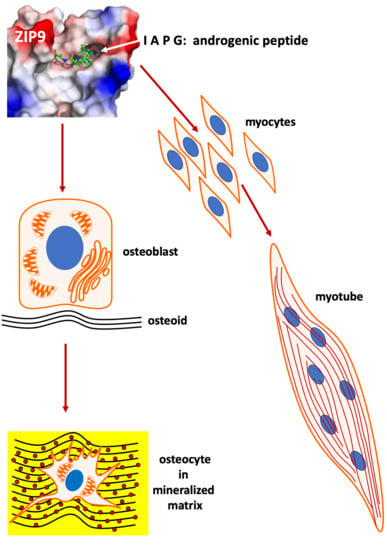
The Effects of Tetrapeptides Designed to Fit the Androgen Binding Site of ZIP9 on Myogenic and Osteogenic Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Modeling of ZIP9 and Docking Experiments to Identify Androgen Binding Sites
2.2. Modeling Peptides within the Androgen Binding Site of ZIP9
2.3. Cell Culture of Rat-Derived Myoblast Cells
2.4. Cell Culture of Human-Derived Osteogenic Cells
2.5. Cell-Surface Labeling with Testosterone-BSA-FITC
2.6. Preparation of Cell Lysates of L6 Myoblasts
2.7. Western Blotting
2.8. Immunofluorescence in Myogenic Cells
2.9. Immunofluorescence in Osteogenic Cells
2.10. Fluorescence Staining, Recording, and Quantification
2.11. Detection of Actin Fibers in L6 Myoblasts
2.12. Alizarin Red Staining and Quantification
2.13. Silencing ZIP9 Expression
2.14. Statistical Analysis
3. Results
3.1. Identification of the Putative Androgen Binding Site of ZIP9 by In Silico Calculations
3.2. Identification of Peptides Fitting the Androgen Binding Site of ZIP9
3.3. Synthesis, Analysis, and Purification of Tetrapeptides
3.4. Binding of Peptides to the Androgen Binding Site of ZIP9
3.5. Physiologically Relevant Responses to Peptides in Myogenic Cells
3.6. Physiologically Relevant Responses in Osteogenic Cells
3.7. Role of ZIP9 in Mineralization Process Induced by Testosterone and Peptides in Osteogenic Cells
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, A.H.; Rice, C.D.; Rahman, M.S.; Dong, J.; Thomas, P. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: I. Discovery in female atlantic croaker and evidence ZIP9 mediates testosterone-induced apoptosis of ovarian follicle cells. Endocrinology 2014, 155, 4237–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shihan, M.; Chan, K.H.; Konrad, L.; Scheiner-Bobis, G. Non-classical testosterone signaling in spermatogenic GC-2 cells is mediated through ZIP9 interacting with Gnalpha11. Cell. Signal. 2015, 27, 2077–2086. [Google Scholar] [CrossRef]
- Thomas, P.; Pang, Y.; Dong, J.; Berg, A.H. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: II. Role of human ZIP9 in testosterone-induced prostate and breast cancer cell apoptosis. Endocrinology 2014, 155, 4250–4265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shihan, M.; Bulldan, A.; Scheiner-Bobis, G. Non-classical testosterone signaling is mediated by a G-protein-coupled receptor interacting with Gnalpha11. Biochim. Biophys. Acta 2014, 1843, 1172–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulldan, A.; Dietze, R.; Shihan, M.; Scheiner-Bobis, G. Non-classical testosterone signaling mediated through ZIP9 stimulates claudin expression and tight junction formation in Sertoli cells. Cell. Signal. 2016, 28, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Bulldan, A.; Malviya, V.N.; Upmanyu, N.; Konrad, L.; Scheiner-Bobis, G. Testosterone/bicalutamide antagonism at the predicted extracellular androgen binding site of ZIP9. Biochim. Biophys. Acta 2017, 1864, 2402–2414. [Google Scholar] [CrossRef] [PubMed]
- Denis, L.J.; Griffiths, K. Endocrine treatment in prostate cancer. Semin. Surg. Oncol. 2000, 18, 52–74. [Google Scholar] [CrossRef]
- Bulldan, A.; Bartsch, J.W.; Konrad, L.; Scheiner-Bobis, G. ZIP9 but not the androgen receptor mediates testosterone-induced migratory activity of metastatic prostate cancer cells. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1857–1868. [Google Scholar] [CrossRef]
- Benten, W.P.; Lieberherr, M.; Giese, G.; Wrehlke, C.; Stamm, O.; Sekeris, C.E.; Mossmann, H.; Wunderlich, F. Functional testosterone receptors in plasma membranes of T cells. FASEB J. 1999, 13, 123–133. [Google Scholar] [CrossRef]
- Gaetjens, E.; Pertschuk, L.P. Synthesis of fluorescein labelled steroid hormone-albumin conjugates for the fluorescent histochemical detection of hormone receptors. J. Steroid Biochem. 1980, 13, 1001–1003. [Google Scholar] [CrossRef]
- Kampa, M.; Papakonstanti, E.A.; Hatzoglou, A.; Stathopoulos, E.N.; Stournaras, C.; Castanas, E. The human prostate cancer cell line LNCaP bears functional membrane testosterone receptors that increase PSA secretion and modify actin cytoskeleton. FASEB J. 2002, 16, 1429–1431. [Google Scholar] [CrossRef]
- Fu, R.; Liu, J.; Fan, J.; Li, R.; Li, D.; Yin, J.; Cui, S. Novel evidence that testosterone promotes cell proliferation and differentiation via G protein-coupled receptors in the rat L6 skeletal muscle myoblast cell line. J. Cell. Physiol. 2012, 227, 98–107. [Google Scholar] [CrossRef]
- Fohr, B.; Schulz, A.; Battmann, A. Sex steroids and bone metabolism: Comparison of in vitro effects of 17beta-estradiol and testosterone on human osteosarcoma cell lines of various gender and differentiation. Exp. Clin. Endocrinol. Diabetes 2000, 108, 414–423. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.; Beer, T.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [Green Version]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef]
- Mackerell, A.D., Jr.; Feig, M.; Brooks, C.L., 3rd. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004, 25, 1400–1415. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Cryst. D Biol. Cryst. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, A.; Tejero, R.; Montelione, G.T. Evaluating protein structures determined by structural genomics consortia. Proteins 2007, 66, 778–795. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Cryst. D Biol. Cryst. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B., 3rd; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, W375–W383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., 3rd; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U.; Luthy, R.; Eisenberg, D. A method to identify protein sequences that fold into a known three-dimensional structure. Science 1991, 253, 164–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luthy, R.; Bowie, J.U.; Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 1992, 356, 83–85. [Google Scholar] [CrossRef]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef] [Green Version]
- Obarska-Kosinska, A.; Iacoangeli, A.; Lepore, R.; Tramontano, A. PepComposer: Computational design of peptides binding to a given protein surface. Nucleic Acids Res. 2016, 44, W522–W528. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [Green Version]
- Duhovny, D.; Nussino, R.; Wolfson, H.J. Efficient Unbound Docking of Rigid Molecules; Rolf Möhring and Rajeev Raman, Ed.; Springer: Rome, Italy, 2002; Volume 2452, pp. 185–200. [Google Scholar]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Boyle, P.M.; Silver, P.A. Harnessing nature’s toolbox: Regulatory elements for synthetic biology. J. R. Soc. Interface 2009, 6, S535–S546. [Google Scholar] [CrossRef] [Green Version]
- Richler, C.; Yaffe, D. The in vitro cultivation and differentiation capacities of myogenic cell lines. Dev. Biol. 1970, 23, 1–22. [Google Scholar] [CrossRef]
- Fogh, J.; Fogh, J.M.; Orfeo, T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 1977, 59, 221–226. [Google Scholar] [CrossRef]
- Mruk, D.D.; Cheng, C.Y. Enhanced chemiluminescence (ECL) for routine immunoblotting: An inexpensive alternative to commercially available kits. Spermatogenesis 2011, 1, 121–122. [Google Scholar] [CrossRef] [Green Version]
- Antala, S.; Ovchinnikov, S.; Kamisetty, H.; Baker, D.; Dempski, R.E. Computation and Functional Studies Provide a Model for the Structure of the Zinc Transporter hZIP4. J. Biol. Chem. 2015, 290, 17796–17805. [Google Scholar] [CrossRef] [Green Version]
- Bafaro, E.M.; Antala, S.; Nguyen, T.V.; Dzul, S.P.; Doyon, B.; Stemmler, T.L.; Dempski, R.E. The large intracellular loop of hZIP4 is an intrinsically disordered zinc binding domain. Metallomics 2015, 7, 1319–1330. [Google Scholar] [CrossRef] [Green Version]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta 2000, 1465, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Faralli, H.; Dilworth, F.J. Turning on myogenin in muscle: A paradigm for understanding mechanisms of tissue-specific gene expression. Comp. Funct. Genom. 2012, 2012, 836374. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.C.; Stewart, C.E.; Sculthorpe, N.; Dugdale, H.F.; Yousefian, F.; Lewis, M.P.; Sharples, A.P. Testosterone enables growth and hypertrophy in fusion impaired myoblasts that display myotube atrophy: Deciphering the role of androgen and IGF-I receptors. Biogerontology 2016, 17, 619–639. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Artaza, J.N.; Taylor, W.E.; Gonzalez-Cadavid, N.F.; Bhasin, S. Androgens stimulate myogenic differentiation and inhibit adipogenesis in C3H 10T1/2 pluripotent cells through an androgen receptor-mediated pathway. Endocrinology 2003, 144, 5081–5088. [Google Scholar] [CrossRef]
- Clemente, C.F.; Corat, M.A.; Saad, S.T.; Franchini, K.G. Differentiation of C2C12 myoblasts is critically regulated by FAK signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005, 289, R862–R870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quach, N.L.; Biressi, S.; Reichardt, L.F.; Keller, C.; Rando, T.A. Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Mol. Biol. Cell 2009, 20, 3422–3435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dabaja, A.A.; Bryson, C.F.; Schlegel, P.N.; Paduch, D.A. The effect of hypogonadism and testosterone-enhancing therapy on alkaline phosphatase and bone mineral density. BJU Int. 2015, 115, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B.L.; Khosla, S. Androgens and bone. Steroids 2009, 74, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.; Hikim, A.P.; Kovacheva, E.L.; Sinha-Hikim, I. Mouse model of testosterone-induced muscle fiber hypertrophy: Involvement of p38 mitogen-activated protein kinase-mediated Notch signaling. J. Endocrinol. 2009, 201, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Connaughton, M.A.; Fine, M.L.; Taylor, M.H. Weakfish sonic muscle: Influence of size, temperature and season. J. Exp. Biol. 2002, 205, 2183–2188. [Google Scholar] [CrossRef]
- Fuxjager, M.J.; Barske, J.; Du, S.; Day, L.B.; Schlinger, B.A. Androgens regulate gene expression in avian skeletal muscles. PLoS ONE 2012, 7, e51482. [Google Scholar] [CrossRef]
- Giangregorio, L.; El-Kotob, R. Exercise, muscle, and the applied load-bone strength balance. Osteoporos. Int. 2017, 28, 21–33. [Google Scholar] [CrossRef]
- Goldspink, G. Loss of muscle strength during aging studied at the gene level. Rejuvenation Res. 2007, 10, 397–405. [Google Scholar] [CrossRef]
- Guadalupe-Grau, A.; Fuentes, T.; Guerra, B.; Calbet, J.A. Exercise and bone mass in adults. Sports Med. 2009, 39, 439–468. [Google Scholar] [CrossRef]
- Sinha-Hikim, I.; Artaza, J.; Woodhouse, L.; Gonzalez-Cadavid, N.; Singh, A.B.; Lee, M.I.; Storer, T.W.; Casaburi, R.; Shen, R.; Bhasin, S. Testosterone-induced increase in muscle size in healthy young men is associated with muscle fiber hypertrophy. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E154–E164. [Google Scholar] [CrossRef]
- Sinha-Hikim, I.; Cornford, M.; Gaytan, H.; Lee, M.L.; Bhasin, S. Effects of testosterone supplementation on skeletal muscle fiber hypertrophy and satellite cells in community-dwelling older men. J. Clin. Endocrinol. Metab. 2006, 91, 3024–3033. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.K. Androgen receptor enhances myogenin expression and accelerates differentiation. Biochem. Biophys. Res. Commun. 2002, 294, 408–413. [Google Scholar] [CrossRef]
- Al-Harbi, T.M.; Bainbridge, L.J.; McQueen, M.J.; Tarnopolsky, M.A. Hypogonadism is common in men with myopathies. J. Clin. Neuromuscul. Dis. 2008, 9, 397–401. [Google Scholar] [CrossRef]
- Wood, C.L.; Cheetham, T.D.; Guglieri, M.; Bushby, K.; Owen, C.; Johnstone, H.; Straub, V. Testosterone Treatment of Pubertal Delay in Duchenne Muscular Dystrophy. Neuropediatrics 2015, 46, 371–376. [Google Scholar] [CrossRef]
- Hegde, S.S.; Revankar, A.V.; Patil, A.K. Identification of bone-specific alkaline phosphatase in saliva and its correlation with skeletal age. Indian. J. Dent. Res. 2018, 29, 721–725. [Google Scholar] [CrossRef]
- Tripathi, T.; Gupta, P.; Sharma, J.; Rai, P.; Gupta, V.K.; Singh, N. Bone-specific alkaline phosphatase—a potential biomarker for skeletal growth assessment. J. Orthod. 2018, 45, 4–10. [Google Scholar] [CrossRef]
- Narayanan, R.; Coss, C.C.; Dalton, J.T. Development of selective androgen receptor modulators (SARMs). Mol. Cell. Endocrinol. 2018, 465, 134–142. [Google Scholar] [CrossRef] [Green Version]








| Template Structure | MolProbity Score | Clash Score | Ramachandran Favoured | Ramachandran Outliers | Rotamer Outliers | C-Beta Deviations | Bad Bonds | Bad Angles |
|---|---|---|---|---|---|---|---|---|
| 5tsa.1.A | 2.61 | 15.74 | 88.01 | 4.79 | 2.59 | 16 | 2/2204 | 67/2999 |
| 6pgi.1.A | 2.48 | 14.66 | 90.03 | 2.75 | 2.16 | 9 | 0/2196 | 40/2988 |
| mouse Zip9 | 2.1 | 0.45 | 85.5 | 5 | 10.9 | 18 | 0/2245 | 40/3054 |
| PepComposer: | ||||||
|---|---|---|---|---|---|---|
| Peptide Sequence | IAPG (Pep-1) | GVSG (Pep-2) | GVVG (Pep-3) | PQTG (Pep-4) | SGNL (Pep-5) | QAPG (Pep-6) |
| FoldX server: | ||||||
| FoldX energy of ZIP9/tetrapeptide complexes (kcal/mol) | −2.2 | −1.8 | −1.2 | −1.2 | −0.8 | −0.5 |
| PatchDock server: | ||||||
| Docking score | 4718 | 4192 | 4082 | 4580 | 4744 | 4744 |
| Surface contact area | 556 | 495 | 461 | 538 | 589 | 524 |
| Atomic contact energy (kJ/mol) | −232 | −215 | −166 | −158 | −201 | −105 |
| AutodockVina: | ||||||
| Binding affinity of peptides for ZIP9 (kcal/mol) | −7.8 | −6.3 | −6.3 | −6.9 | −6.3 | −7.7 |
| Testosterone | IAPG (Pep-1) | GVSG (Pep-2) | GVVG (Pep-3) | PQTG (Pep-4) | SGNL (Pep-5) | QAPG (Pep-6) |
|---|---|---|---|---|---|---|
| A165 | ||||||
| A166 | A166 | A166 | A166 | A166 | A166 | A166 |
| S170 | S170 | S170 | S170 | S170 | ||
| S173 | S173 | |||||
| V174 | V174 | |||||
| A181 | ||||||
| S230 | S230 | S230 | S230 | S230 | ||
| S232 | S232 | S232 | ||||
| S233 | S233 | |||||
| V240 | V240 | V240 | ||||
| M247 | M247 | M247 | ||||
| L248 | L248 | L248 | ||||
| G244 | G244 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malviya, V.N.; Bulldan, A.; Wende, R.C.; Kabbesh, H.; Möller, M.-L.; Schreiner, P.R.; Scheiner-Bobis, G. The Effects of Tetrapeptides Designed to Fit the Androgen Binding Site of ZIP9 on Myogenic and Osteogenic Cells. Biology 2022, 11, 19. https://doi.org/10.3390/biology11010019
Malviya VN, Bulldan A, Wende RC, Kabbesh H, Möller M-L, Schreiner PR, Scheiner-Bobis G. The Effects of Tetrapeptides Designed to Fit the Androgen Binding Site of ZIP9 on Myogenic and Osteogenic Cells. Biology. 2022; 11(1):19. https://doi.org/10.3390/biology11010019
Chicago/Turabian StyleMalviya, Viveka Nand, Ahmed Bulldan, Raffael Christoph Wende, Hassan Kabbesh, Marie-Louise Möller, Peter Richard Schreiner, and Georgios Scheiner-Bobis. 2022. "The Effects of Tetrapeptides Designed to Fit the Androgen Binding Site of ZIP9 on Myogenic and Osteogenic Cells" Biology 11, no. 1: 19. https://doi.org/10.3390/biology11010019
APA StyleMalviya, V. N., Bulldan, A., Wende, R. C., Kabbesh, H., Möller, M. -L., Schreiner, P. R., & Scheiner-Bobis, G. (2022). The Effects of Tetrapeptides Designed to Fit the Androgen Binding Site of ZIP9 on Myogenic and Osteogenic Cells. Biology, 11(1), 19. https://doi.org/10.3390/biology11010019