The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield
Abstract
:Simple Summary
Abstract
1. Introduction
2. Strigolactone Classification and Synthesis
3. Strigolactone Signalling Mechanism
4. Strigolactone-Mediated Bud Outgrowth
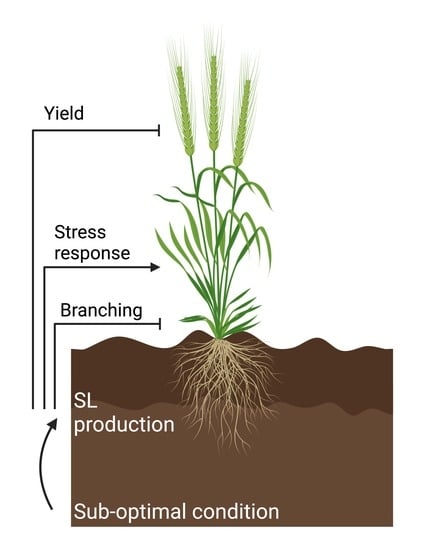
5. Strigolactone-Mediated Responses to Sub-Optimal Conditions
6. Targeting the Strigolactone Pathway for Crop Improvement
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef] [PubMed]
- Saeed, W.; Naseem, S.; Ali, Z. Strigolactones Biosynthesis and Their Role in Abiotic Stress Resilience in Plants: A Critical Review. Front Plant Sci. 2017, 8, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Mazur, E.; Balla, J.; Gallei, M.; Kalousek, P.; Medvedova, Z.; Li, Y.; Wang, Y.; Prat, T.; Vasileva, M.; et al. Strigolactones inhibit auxin feedback on PIN-dependent auxin transport canalization. Nat. Commun. 2020, 11, 3508. [Google Scholar] [CrossRef]
- Studer, A.; Zhao, Q.; Ross-Ibarra, J.; Doebley, J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 2011, 43, 1160–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, H.; Jiang, Z.; Wang, W.; Xu, R.; Wang, Q.; Zhang, Z.; Li, A.; Liang, Y.; Ou, S.; et al. Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 2021, 590, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Cook, C.E.; Whichard, L.P.; Turner, B.; Wall, M.E.; Egley, G.H. Germination of Witchweed (Striga lutea Lour.): Isolation and Properties of a Potent Stimulant. Science 1966, 154, 1189–1190. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Brewer, P.B. Strigolactones, how are they synthesized to regulate plant growth and development? Curr. Opin. Plant Biol. 2021, 63, 102072. [Google Scholar] [CrossRef]
- Lopez-Obando, M.; Ligerot, Y.; Bonhomme, S.; Boyer, F.D.; Rameau, C. Strigolactone biosynthesis and signaling in plant development. Development 2015, 142, 3615–3619. [Google Scholar] [CrossRef] [Green Version]
- Yokota, T.; Sakai, H.; Okuno, K.; Yoneyama, K.; Takeuchi, Y. Alectrol and orobanchol, germination stimulants for Orobanche minor, from its host red clover. Phytochemistry 1998, 49, 1967–1973. [Google Scholar] [CrossRef]
- Zwanenburg, B.; Pospisil, T. Structure and activity of strigolactones: New plant hormones with a rich future. Mol. Plant 2013, 6, 38–62. [Google Scholar] [CrossRef]
- Yoneyama, K. Recent progress in the chemistry and biochemistry of strigolactones. J. Pestic. Sci. 2020, 45, 45–53. [Google Scholar] [CrossRef] [Green Version]
- Alder, A.; Jamil, M.; Marzorati, M.; Bruno, M.; Vermathen, M.; Bigler, P.; Ghisla, S.; Bouwmeester, H.; Beyer, P.; Al-Babili, S. The path from beta-carotene to carlactone, a strigolactone-like plant hormone. Science 2012, 335, 1348–1351. [Google Scholar] [CrossRef] [Green Version]
- Booker, J.; Auldridge, M.; Wills, S.; McCarty, D.; Klee, H.; Leyser, O. MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr. Biol. 2004, 14, 1232–1238. [Google Scholar] [CrossRef] [Green Version]
- Sorefan, K.; Booker, J.; Haurogne, K.; Goussot, M.; Bainbridge, K.; Foo, E.; Chatfield, S.; Ward, S.; Beveridge, C.; Rameau, C.; et al. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes Dev. 2003, 17, 1469–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, Y.; Sado, A.; Asami, K.; Hanada, A.; Umehara, M.; Akiyama, K.; Yamaguchi, S. Carlactone is an endogenous biosynthetic precursor for strigolactones. Proc. Natl. Acad. Sci. USA 2014, 111, 1640–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Booker, J.; Sieberer, T.; Wright, W.; Williamson, L.; Willett, B.; Stirnberg, P.; Turnbull, C.; Srinivasan, M.; Goddard, P.; Leyser, O. MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid-derived branch-inhibiting hormone. Dev. Cell 2005, 8, 443–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abe, S.; Sado, A.; Tanaka, K.; Kisugi, T.; Asami, K.; Ota, S.; Kim, H.I.; Yoneyama, K.; Xie, X.; Ohnishi, T.; et al. Carlactone is converted to carlactonoic acid by MAX1 in Arabidopsis and its methyl ester can directly interact with AtD14 in vitro. Proc. Natl. Acad. Sci. USA 2014, 111, 18084–18089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneyama, K.; Mori, N.; Sato, T.; Yoda, A.; Xie, X.; Okamoto, M.; Iwanaga, M.; Ohnishi, T.; Nishiwaki, H.; Asami, T.; et al. Conversion of carlactone to carlactonoic acid is a conserved function of MAX1 homologs in strigolactone biosynthesis. New Phytol. 2018, 218, 1522–1533. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Yoneyama, K.; Kisugi, T.; Uchida, K.; Ito, S.; Akiyama, K.; Hayashi, H.; Yokota, T.; Nomura, T.; Yoneyama, K. Confirming stereochemical structures of strigolactones produced by rice and tobacco. Mol. Plant 2013, 6, 153–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashiguchi, K.; Seto, Y.; Onozuka, Y.; Suzuki, S.; Takemoto, K.; Wang, Y.; Dong, L.; Asami, K.; Noda, R.; Kisugi, T.; et al. A carlactonoic acid methyltransferase that contributes to the inhibition of shoot branching in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2111565119. [Google Scholar] [CrossRef]
- Brewer, P.B.; Yoneyama, K.; Filardo, F.; Meyers, E.; Scaffidi, A.; Frickey, T.; Akiyama, K.; Seto, Y.; Dun, E.A.; Cremer, J.E.; et al. LATERAL BRANCHING OXIDOREDUCTASE acts in the final stages of strigolactone biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6301–6306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, E.; Chai, L.; Zhang, S.; Yu, R.; Zhang, X.; Xu, C.; Hu, Y. Catabolism of strigolactones by a carboxylesterase. Nat. Plants 2021, 7, 1495–1504. [Google Scholar] [CrossRef]
- Roesler, K.; Lu, C.; Thomas, J.; Xu, Q.; Vance, P.; Hou, Z.; Williams, R.W.; Liu, L.; Owens, M.A.; Habben, J.E. Arabidopsis Carboxylesterase 20 Binds Strigolactone and Increases Branches and Tillers When Ectopically Expressed in Arabidopsis and Maize. Front. Plant Sci. 2021, 12, 639401. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Situmorang, A.; Brewer, P.B.; Braszewska, A. Diverse Roles of MAX1 Homologues in Rice. Genes 2020, 11, 1348. [Google Scholar] [CrossRef] [PubMed]
- Shiratake, K.; Notaguchi, M.; Makino, H.; Sawai, Y.; Borghi, L. Petunia PLEIOTROPIC DRUG RESISTANCE 1 is a Strigolactone Short-Distance Transporter with Long-Distance Outcomes. Plant Cell Physiol. 2019, 60, 1722–1733. [Google Scholar] [CrossRef]
- Ito, S.; Braguy, J.; Wang, J.Y.; Yoda, A.; Fiorilli, V.; Takahashi, I.; Jamil, M.; Felemban, A.; Miyazaki, S.; Mazzarella, T.; et al. Canonical strigolactones are not the major determinant of tillering but important rhizospheric signals in rice. Sci. Adv. 2022, 8, eadd1278. [Google Scholar] [CrossRef] [PubMed]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef] [Green Version]
- Waters, M.T.; Nelson, D.C.; Scaffidi, A.; Flematti, G.R.; Sun, Y.K.; Dixon, K.W.; Smith, S.M. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 2012, 139, 1285–1295. [Google Scholar] [CrossRef] [Green Version]
- Hamiaux, C.; Drummond, R.S.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an alpha/beta hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef] [Green Version]
- Marzec, M.; Gruszka, D.; Tylec, P.; Szarejko, I. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant 2016, 158, 341–355. [Google Scholar] [CrossRef]
- Stirnberg, P.; Furner, I.J.; Ottoline Leyser, H.M. MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 2007, 50, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Lin, Q.; Zhu, L.; Ren, Y.; Zhou, K.; Shabek, N.; Wu, F.; Mao, H.; Dong, W.; Gan, L.; et al. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 2013, 504, 406–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Liu, X.; Xiong, G.; Liu, H.; Chen, F.; Wang, L.; Meng, X.; Liu, G.; Yu, H.; Yuan, Y.; et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 2013, 504, 401–405. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, B.; Yu, H.; Guo, H.; Lin, T.; Kou, L.; Wang, A.; Shao, N.; Ma, H.; Xiong, G.; et al. Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 2020, 583, 277–281. [Google Scholar] [CrossRef]
- Waters, M.T.; Gutjahr, C.; Bennett, T.; Nelson, D.C. Strigolactone Signaling and Evolution. Annu. Rev. Plant Biol. 2017, 68, 291–322. [Google Scholar] [CrossRef]
- Yao, R.; Ming, Z.; Yan, L.; Li, S.; Wang, F.; Ma, S.; Yu, C.; Yang, M.; Chen, L.; Chen, L.; et al. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 2016, 536, 469–473. [Google Scholar] [CrossRef]
- Marzec, M.; Brewer, P. Binding or Hydrolysis? How Does the Strigolactone Receptor Work? Trends Plant Sci. 2019, 24, 571–574. [Google Scholar] [CrossRef]
- Shabek, N.; Ticchiarelli, F.; Mao, H.; Hinds, T.R.; Leyser, O.; Zheng, N. Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling. Nature 2018, 563, 652–656. [Google Scholar] [CrossRef]
- Li, Q.; Martin-Fontecha, E.S.; Khosla, A.; White, A.R.F.; Chang, S.; Cubas, P.; Nelson, D.C. The strigolactone receptor D14 targets SMAX1 for degradation in response to GR24 treatment and osmotic stress. Plant Commun. 2022, 3, 100303. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Sun, H.; Li, W.; Burritt, D.J.; Tian, H.; Zhang, H.; Liang, X.; Miao, Y.; Mostofa, M.G.; Tran, L.-S.P. Strigolactones interact with other phytohormones to modulate plant root growth and development. Crop J. 2022, 10, 1517–1527. [Google Scholar] [CrossRef]
- Kaniganti, S.; Bhattacharya, J.; Petla, B.P.; Reddy, P.S. Strigolactone, a neglected plant hormone, with a great potential for crop improvement: Crosstalk with other plant hormones. Environ. Exp. Bot. 2022, 204, 105072. [Google Scholar] [CrossRef]
- Bennett, T.; Hines, G.; van Rongen, M.; Waldie, T.; Sawchuk, M.G.; Scarpella, E.; Ljung, K.; Leyser, O. Connective Auxin Transport in the Shoot Facilitates Communication between Shoot Apices. PLoS Biol. 2016, 14, e1002446. [Google Scholar] [CrossRef] [Green Version]
- Thimann, K.V.; Skoog, F. Studies on the Growth Hormone of Plants: III. The Inhibiting Action of the Growth Substance on Bud Development. Proc. Natl. Acad. Sci. USA 1933, 19, 714–716. [Google Scholar] [CrossRef] [Green Version]
- Hall, S.M.; Hillman, J.R. Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. timing of bud growth following decapitation. Planta 1975, 123, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Wisniewska, J.; Xu, J.; Seifertova, D.; Brewer, P.B.; Ruzicka, K.; Blilou, I.; Rouquie, D.; Benkova, E.; Scheres, B.; Friml, J. Polar PIN localization directs auxin flow in plants. Science 2006, 312, 883. [Google Scholar] [CrossRef] [Green Version]
- Balla, J.; Medvedova, Z.; Kalousek, P.; Matijescukova, N.; Friml, J.; Reinohl, V.; Prochazka, S. Auxin flow-mediated competition between axillary buds to restore apical dominance. Sci. Rep. 2016, 6, 35955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chabikwa, T.G.; Brewer, P.B.; Beveridge, C.A. Initial Bud Outgrowth Occurs Independent of Auxin Flow from Out of Buds. Plant Physiol. 2019, 179, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Bennett, T.; Sieberer, T.; Willett, B.; Booker, J.; Luschnig, C.; Leyser, O. The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 2006, 16, 553–563. [Google Scholar] [CrossRef]
- Crawford, S.; Shinohara, N.; Sieberer, T.; Williamson, L.; George, G.; Hepworth, J.; Muller, D.; Domagalska, M.A.; Leyser, O. Strigolactones enhance competition between shoot branches by dampening auxin transport. Development 2010, 137, 2905–2913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009, 150, 482–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brewer, P.B.; Dun, E.A.; Gui, R.; Mason, M.G.; Beveridge, C.A. Strigolactone Inhibition of Branching Independent of Polar Auxin Transport. Plant Physiol. 2015, 168, 1820–1829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguilar-Martinez, J.A.; Poza-Carrion, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Dun, E.A.; de Saint Germain, A.; Rameau, C.; Beveridge, C.A. Antagonistic action of strigolactone and cytokinin in bud outgrowth control. Plant Physiol. 2012, 158, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Braun, N.; de Saint Germain, A.; Pillot, J.P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Waldie, T.; Leyser, O. Cytokinin Targets Auxin Transport to Promote Shoot Branching. Plant Physiol. 2018, 177, 803–818. [Google Scholar] [CrossRef] [Green Version]
- Choubane, D.; Rabot, A.; Mortreau, E.; Legourrierec, J.; Peron, T.; Foucher, F.; Ahcene, Y.; Pelleschi-Travier, S.; Leduc, N.; Hamama, L.; et al. Photocontrol of bud burst involves gibberellin biosynthesis in Rosa sp. J. Plant Physiol. 2012, 169, 1271–1280. [Google Scholar] [CrossRef]
- Ito, S.; Yamagami, D.; Umehara, M.; Hanada, A.; Yoshida, S.; Sasaki, Y.; Yajima, S.; Kyozuka, J.; Ueguchi-Tanaka, M.; Matsuoka, M.; et al. Regulation of Strigolactone Biosynthesis by Gibberellin Signaling. Plant Physiol. 2017, 174, 1250–1259. [Google Scholar] [CrossRef]
- Ni, J.; Gao, C.; Chen, M.S.; Pan, B.Z.; Ye, K.; Xu, Z.F. Gibberellin Promotes Shoot Branching in the Perennial Woody Plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef]
- Gonzalez-Grandio, E.; Poza-Carrion, C.; Sorzano, C.O.; Cubas, P. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. Plant Cell 2013, 25, 834–850. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Le Moigne, M.A.; Bertheloot, J.; Crespel, L.; Perez-Garcia, M.D.; Oge, L.; Demotes-Mainard, S.; Hamama, L.; Daviere, J.M.; Sakr, S. BRANCHED1: A Key Hub of Shoot Branching. Front. Plant Sci. 2019, 10, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kebrom, T.H. A Growing Stem Inhibits Bud Outgrowth—The Overlooked Theory of Apical Dominance. Front. Plant Sci. 2017, 8, 1874. [Google Scholar] [CrossRef] [Green Version]
- Mostofa, M.G.; Li, W.; Nguyen, K.H.; Fujita, M.; Tran, L.P. Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell Environ. 2018, 41, 2227–2243. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Magome, H.; Takeda-Kamiya, N.; Yamaguchi, S. Contribution of strigolactones to the inhibition of tiller bud outgrowth under phosphate deficiency in rice. Plant Cell Physiol. 2010, 51, 1118–1126. [Google Scholar] [CrossRef] [Green Version]
- Wu, K.; Wang, S.; Song, W.; Zhang, J.; Wang, Y.; Liu, Q.; Yu, J.; Ye, Y.; Li, S.; Chen, J.; et al. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice. Science 2020, 367, eaaz2046. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Song, X.; Lu, Z.; Yu, H.; Shao, G.; Xiong, J.; Meng, X.; Jing, Y.; Liu, G.; Xiong, G.; Duan, J.; et al. IPA1 functions as a downstream transcription factor repressed by D53 in strigolactone signaling in rice. Cell Res. 2017, 27, 1128–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Guo, X.; Qi, X.; Feng, F.; Xie, X.; Zhang, Y.; Zhao, Q. SPL14/17 act downstream of strigolactone signalling to modulate rice root elongation in response to nitrate supply. Plant J. 2021, 106, 649–660. [Google Scholar] [CrossRef]
- Marzec, M.; Daszkowska-Golec, A.; Collin, A.; Melzer, M.; Eggert, K.; Szarejko, I. Barley strigolactone signalling mutant hvd14.d reveals the role of strigolactones in abscisic acid-dependent response to drought. Plant Cell Environ. 2020, 43, 2239–2253. [Google Scholar] [CrossRef]
- Ha, C.V.; Leyva-Gonzalez, M.A.; Osakabe, Y.; Tran, U.T.; Nishiyama, R.; Watanabe, Y.; Tanaka, M.; Seki, M.; Yamaguchi, S.; Dong, N.V.; et al. Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. USA 2014, 111, 851–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; He, H.; Vitali, M.; Visentin, I.; Charnikhova, T.; Haider, I.; Schubert, A.; Ruyter-Spira, C.; Bouwmeester, H.J.; Lovisolo, C.; et al. Osmotic stress represses strigolactone biosynthesis in Lotus japonicus roots: Exploring the interaction between strigolactones and ABA under abiotic stress. Planta 2015, 241, 1435–1451. [Google Scholar] [CrossRef] [PubMed]
- Brun, G. At the crossroads of strigolactones and abscisic acid pathways: A role for miR156. Plant Cell Environ. 2020, 43, 1609–1612. [Google Scholar] [CrossRef]
- Yao, C.; Finlayson, S.A. Abscisic Acid is a General Negative Regulator of Arabidopsis Axillary Bud Growth. Plant Physiol. 2015, 169, 611–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.J.; Luo, J.; Wang, Y.; Li, N. From Green Revolution to Green Balance: The Nitrogen and Gibberellin Mediated Rice Tiller Growth. Plant Signal. Behav. 2021, 16, 1917838. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hochman, Z.; Horan, H. Causes of wheat yield gaps and opportunities to advance the water-limited yield frontier in Australia. Field Crops Res. 2018, 228, 20–30. [Google Scholar] [CrossRef]
- Menegat, S.; Ledo, A.; Tirado, R. Greenhouse gas emissions from global production and use of nitrogen synthetic fertilisers in agriculture. Sci. Rep. 2022, 12, 14490. [Google Scholar] [CrossRef]
- Smith, M.R.; Rao, I.M.; Merchant, A. Source-Sink Relationships in Crop Plants and Their Influence on Yield Development and Nutritional Quality. Front. Plant Sci. 2018, 9, 1889. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Jang, S.; Lee, Y.K.; Kim, D.G.; Jin, Z.; Koh, H.J. Genetic Basis of Tiller Dynamics of Rice Revealed by Genome-Wide Association Studies. Plants 2020, 9, 1695. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, L.; Yu, H.; Zeng, L.; Hu, J.; Ni, S.; Rao, Y.; Li, S.; Chu, J.; Meng, X.; et al. A Strigolactone Biosynthesis Gene Contributed to the Green Revolution in Rice. Mol. Plant 2020, 13, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Alvi, A.F.; Sehar, Z.; Fatma, M.; Masood, A.; Khan, N.A. Strigolactone: An Emerging Growth Regulator for Developing Resilience in Plants. Plants 2022, 11, 2604. [Google Scholar] [CrossRef] [PubMed]
- Jamil, M.; Wang, J.Y.; Yonli, D.; Patil, R.H.; Riyazaddin, M.; Gangashetty, P.; Berqdar, L.; Chen, G.E.; Traore, H.; Margueritte, O.; et al. A New Formulation for Strigolactone Suicidal Germination Agents, towards Successful Striga Management. Plants 2022, 11, 808. [Google Scholar] [CrossRef] [PubMed]
- Cartry, D.; Steinberg, C.; Gibot-Leclerc, S. Main drivers of broomrape regulation. A review. Agron. Sustain. Dev. 2021, 41, 17. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, H.; Ma, B.; Liu, G.; Wang, J.; Wang, J.; Gao, R.; Li, J.; Liu, J.; Xu, J.; et al. A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 2017, 8, 14789. [Google Scholar] [CrossRef] [Green Version]
- Ren, M.; Huang, M.; Qiu, H.; Chun, Y.; Li, L.; Kumar, A.; Fang, J.; Zhao, J.; He, H.; Li, X. Genome-Wide Association Study of the Genetic Basis of Effective Tiller Number in Rice. Rice 2021, 14, 56. [Google Scholar] [CrossRef]
- Bajic, M.; Maher, K.A.; Deal, R.B. Identification of Open Chromatin Regions in Plant Genomes Using ATAC-Seq. Methods Mol. Biol. 2018, 1675, 183–201. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Gallagher, J.; Arevalo, E.D.; Chen, R.; Skopelitis, T.; Wu, Q.; Bartlett, M.; Jackson, D. Enhancing grain-yield-related traits by CRISPR-Cas9 promoter editing of maize CLE genes. Nat. Plants 2021, 7, 287–294. [Google Scholar] [CrossRef]
- Guan, J.C.; Li, C.; Flint-Garcia, S.; Suzuki, M.; Wu, S.; Saunders, J.W.; Dong, L.; Bouwmeester, H.J.; McCarty, D.R.; Koch, K.E. Maize domestication phenotypes reveal strigolactone networks coordinating grain size evolution with kernel-bearing cupule architecture. Plant Cell 2022. [Google Scholar] [CrossRef]
- Liu, R.; Hou, J.; Li, H.; Xu, P.; Zhang, Z.; Zhang, X. Association of TaD14-4D, a Gene Involved in Strigolactone Signaling, with Yield Contributing Traits in Wheat. Int. J. Mol. Sci. 2021, 22, 3748. [Google Scholar] [CrossRef]
- Yao, F.Q.; Li, X.H.; Wang, H.; Song, Y.N.; Li, Z.Q.; Li, X.G.; Gao, X.Q.; Zhang, X.S.; Bie, X.M. Down-expression of TaPIN1s Increases the Tiller Number and Grain Yield in Wheat. BMC Plant Biol. 2021, 21, 443. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, J.H.; Tucker, M.R.; Brewer, P.B. The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield. Biology 2023, 12, 95. https://doi.org/10.3390/biology12010095
Kelly JH, Tucker MR, Brewer PB. The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield. Biology. 2023; 12(1):95. https://doi.org/10.3390/biology12010095
Chicago/Turabian StyleKelly, Jack H., Matthew R. Tucker, and Philip B. Brewer. 2023. "The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield" Biology 12, no. 1: 95. https://doi.org/10.3390/biology12010095
APA StyleKelly, J. H., Tucker, M. R., & Brewer, P. B. (2023). The Strigolactone Pathway Is a Target for Modifying Crop Shoot Architecture and Yield. Biology, 12(1), 95. https://doi.org/10.3390/biology12010095