A Multi-Functional Tubulovesicular Network as the Ancestral Eukaryotic Endomembrane System
Abstract
:1. Introduction
2. Literature Review
2.1 The Combination of the Organelle Paralogy and Protocoatomer Hypotheses Explains Autogenous Organelle Evolution
2.1.1. The Organelle Paralogy Hypothesis
2.1.2. The Protocoatomer Hypothesis

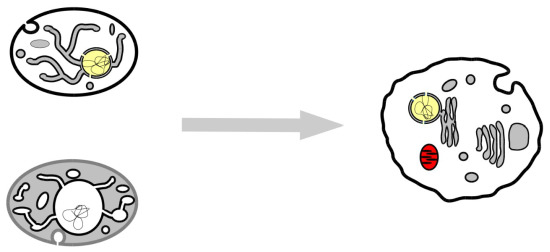
3. Our Proposal
3.1. Both OPH and PCH Predict an Ancestral Eukaryotic ES with Simpler Organisation, Fewer Proteins and Less Specialized Function
3.2. What Shape Could the Ancestral Eukaryotic ES Have Presented?
3.3. Tubulovesicular Network in Modern Organisms

3.4. What Function Could the Ancestral Eukaryotic ES Have Contained?
Function of Current TVNs
3.5. A Coherent Model of Complex Intracellular Membrane System Evolution

3.6. Current Organisms Illustrate Our Scenario
| Steps | Name | Features | Examples | Illustrations |
|---|---|---|---|---|
| 1 | Simple | None (monoderm or diderm) | Prokaryotes (most of them, both archaea and bacteria) |  |
| 2 | Vesicles | Vesicle or Saccules | Magnetotactic; photosynthetic and anammox bacteria |  |
| 3 | Invaginations | IM invaginations | PVC bacteria |  |
| 4 | TVN | Connected tubules and vesicles | G. obscuriglobus (B), Ignicoccus hospitalis (A), and Giardia lamblia (E) |  |
| 5 | Developed | Functionally specialized and spatially separated | Eukaryotes (most of them) |  |
4. Discussion
5. Conclusions
Acknowledgements
Author Contributions
Abbreviations
| ES | Endomembrane system |
| OPH | organelles paralogy hypothesis |
| PCH | protocoatomer hypothesis |
| MC | membrane coat |
| TVN | tubulovesicular network |
Conflicts of Interest
References
- Koumandou, V.L.; Wickstead, B.; Ginger, M.L.; van der Giezen, M.; Dacks, J.B.; Field, M.C. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 373–396. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. The origin of eukaryotic and archaebacterial cells. Ann. N. Y. Acad. Sci. 1987, 503, 17–54. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Mentel, M.; van Hellemond, J.J.; Henze, K.; Woehle, C.; Gould, S.B.; Yu, R.-Y.; van der Giezen, M.; Tielens, A.G.M.; Martin, W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012, 76, 444–495. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J. The endosymbiotic origin, diversification and fate of plastids. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 729–748. [Google Scholar] [CrossRef]
- Wideman, J.G.; Leung, K.F.; Field, M.C.; Dacks, J.B. The cell biology of the endocytic system from an evolutionary perspective. Cold Spring Harb. Perspect. Biol. 2014, 6, a016998. [Google Scholar] [CrossRef] [PubMed]
- Elias, M.; Brighouse, A.; Gabernet-Castello, C.; Field, M.C.; Dacks, J.B. Sculpting the endomembrane system in deep time: High resolution phylogenetics of Rab GTPases. J. Cell Sci. 2012, 125, 2500–2508. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.; Dokudovskaya, S.; Alber, F.; Williams, R.; Chait, B.T.; Sali, A.; Rout, M.P. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLOS Biol. 2004, 2, e380. [Google Scholar] [CrossRef] [PubMed]
- Dacks, J.B.; Poon, P.P.; Field, M.C. Phylogeny of endocytic components yields insight into the process of nonendosymbiotic organelle evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Wideman, J.G.; Gawryluk, R.M.R.; Gray, M.W.; Dacks, J.B. The ancient and widespread nature of the ER-mitochondria encounter structure. Mol. Biol. Evol. 2013, 30, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Glick, B.S. The mechanisms of vesicle budding and fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Schledzewski, K.; Brinkmann, H.; Mendel, R.R. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J. Mol. Evol. 1999, 48, 770–778. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, Y.; Seixas, E.; Gouw, M.; Tavares-Cadete, F.; Seabra, M.C.; Pereira-Leal, J.B. Thousands of rab GTPases for the cell biologist. PLOS Comput. Biol. 2011, 7, e1002217. [Google Scholar] [CrossRef] [PubMed]
- Boehm, M.; Bonifacino, J.S. Adaptins the final recount. Mol. Biol. Cell 2001, 12, 2907–2920. [Google Scholar] [CrossRef] [PubMed]
- Carvalho-Santos, Z.; Azimzadeh, J.; Pereira-Leal, J.B.; Bettencourt-Dias, M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011, 194, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Dacks, J.B.; Doolittle, W.F. Novel syntaxin gene sequences from Giardia, Trypanosoma and algae: Implications for the ancient evolution of the eukaryotic endomembrane system. J. Cell Sci. 2002, 115, 1635–1642. [Google Scholar] [PubMed]
- Dacks, J.B.; Doolittle, W.F. Molecular and phylogenetic characterization of syntaxin genes from parasitic protozoa. Mol. Biochem. Parasitol. 2004, 136, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Schlacht, A.; Mowbrey, K.; Elias, M.; Kahn, R.A.; Dacks, J.B. Ancient complexity, opisthokont plasticity, and discovery of the 11th subfamily of Arf GAP proteins. Traffic Cph. Den. 2013, 14, 636–649. [Google Scholar] [CrossRef]
- Dacks, J.B.; Peden, A.A.; Field, M.C. Evolution of specificity in the eukaryotic endomembrane system. Int. J. Biochem. Cell Biol. 2009, 41, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Dacks, J.B.; Field, M.C. Evolution of the eukaryotic membrane-trafficking system: Origin, tempo and mode. J. Cell Sci. 2007, 120, 2977–2985. [Google Scholar] [CrossRef] [PubMed]
- Ramadas, R.; Thattai, M. New organelles by gene duplication in a biophysical model of eukaryote endomembrane evolution. Biophys. J. 2013, 104, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Dokudovskaya, S.; Waharte, F.; Schlessinger, A.; Pieper, U.; Devos, D.P.; Cristea, I.M.; Williams, R.; Salamero, J.; Chait, B.T.; Sali, A.; et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics 2011. [Google Scholar] [CrossRef]
- Van Dam, T.J.P.; Townsend, M.J.; Turk, M.; Schlessinger, A.; Sali, A.; Field, M.C.; Huynen, M.A. Evolution of modular intraflagellar transport from a coatomer-like progenitor. Proc. Natl. Acad. Sci. USA 2013, 110, 6943–6948. [Google Scholar] [CrossRef] [PubMed]
- Balderhaar, H.J.; Ungermann, C. CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 2013, 126, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Hirst, J.; Schlacht, A.; Norcott, J.P.; Traynor, D.; Bloomfield, G.; Antrobus, R.; Kay, R.R.; Dacks, J.B.; Robinson, M.S. Characterization of TSET, an ancient and widespread membrane trafficking complex. eLife 2014, 3, e02866. [Google Scholar] [CrossRef] [PubMed]
- Field, M.C.; Sali, A.; Rout, M.P. Evolution: On a bender—BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 2011, 193, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Field, M.; Koreny, L.; Rout, M. Enriching the pore: Splendid complexity from humble origins. Traffic 2014, in press. [Google Scholar]
- Devos, D.P. Gemmata obscuriglobus. Curr. Biol. 2013, 23, R705–R707. [Google Scholar] [CrossRef] [PubMed]
- Fuerst, J.A. Intracellular compartmentation in planctomycetes. Annu. Rev. Microbiol. 2005, 59, 299–328. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.P. PVC bacteria: Variation of, but not exception to, the Gram-negative cell plan. Trends Microbiol. 2014, 22, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, M.R.; Webb, R.I.; Strous, M.; Jetten, M.S.; Butler, M.K.; Forde, R.J.; Fuerst, J.A. Cell compartmentalisation in planctomycetes: Novel types of structural organisation for the bacterial cell. Arch. Microbiol. 2001, 175, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Jogler, C.; Glöckner, F.O.; Kolter, R. Characterization of Planctomyces limnophilus and development of genetic tools for its manipulation establish it as a model species for the phylum Planctomycetes. Appl. Environ. Microbiol. 2011, 77, 5826–5829. [Google Scholar] [CrossRef] [PubMed]
- Lieber, A.; Leis, A.; Kushmaro, A.; Minsky, A.; Medalia, O. Chromatin organization and radio resistance in the bacterium gemmata obscuriglobus. J. Bacteriol. 2009, 191, 1439–1445. [Google Scholar] [CrossRef] [PubMed]
- Santarella-Mellwig, R.; Pruggnaller, S.; Roos, N.; Mattaj, I.W.; Devos, D.P. Three-dimensional reconstruction of bacteria with a complex endomembrane system. PLOS Biol. 2013, 11, e1001565. [Google Scholar] [CrossRef] [PubMed]
- Acehan, D.; Santarella-Mellwig, R.; Devos, D.P. A bacterial tubulovesicular network. J. Cell Sci. 2014, 127, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Huber, H.; Hohn, M.J.; Rachel, R.; Fuchs, T.; Wimmer, V.C.; Stetter, K.O. A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature 2002, 417, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Rachel, R.; Wyschkony, I.; Riehl, S.; Huber, H. The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon. Archaea Vanc. BC 2002, 1, 9–18. [Google Scholar] [CrossRef]
- Junglas, B.; Briegel, A.; Burghardt, T.; Walther, P.; Wirth, R.; Huber, H.; Rachel, R. Ignicoccus hospitalis and Nanoarchaeum equitans: Ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch. Microbiol. 2008, 190, 395–408. [Google Scholar] [CrossRef] [PubMed]
- Dridi, B.; Fardeau, M.-L.; Ollivier, B.; Raoult, D.; Drancourt, M. Methanomassiliicoccus luminyensis gen. nov., sp. nov., a methanogenic archaeon isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2012, 62, 1902–1907. [Google Scholar] [CrossRef] [PubMed]
- Beznoussenko, G.V.; Dolgikh, V.V.; Seliverstova, E.V.; Semenov, P.B.; Tokarev, Y.S.; Trucco, A.; Micaroni, M.; di Giandomenico, D.; Auinger, P.; Senderskiy, I.V.; et al. Analogs of the Golgi complex in microsporidia: Structure and avesicular mechanisms of function. J. Cell Sci. 2007, 120, 1288–1298. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.E.; Huston, C.D. Evidence of a continuous endoplasmic reticulum in the protozoan parasite Entamoeba histolytica. Eukaryot. Cell 2008, 7, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Vaithilingam, A.; Teixeira, J.E.; Huston, C.D. Endoplasmic reticulum continuity in the protozoan parasite Entamoeba histolytica. Commun. Integr. Biol. 2008, 1, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Abodeely, M.; DuBois, K.N.; Hehl, A.; Stefanic, S.; Sajid, M.; DeSouza, W.; Attias, M.; Engel, J.C.; Hsieh, I.; Fetter, R.D.; et al. A contiguous compartment functions as endoplasmic reticulum and endosome/lysosome in Giardia lamblia. Eukaryot. Cell 2009, 8, 1665–1676. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; Roger, A.J. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 1995, 92, 6518–6521. [Google Scholar] [CrossRef] [PubMed]
- Santarella-Mellwig, R.; Franke, J.; Jaedicke, A.; Gorjanacz, M.; Bauer, U.; Budd, A.; Mattaj, I.W.; Devos, D.P. The compartmentalized bacteria of the planctomycetes-verrucomicrobia-chlamydiae superphylum have membrane coat-like proteins. PLOS Biol. 2010, 8, e1000281. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.P.; Andalusian Center for Developmental Biology (CABD), UPO, Sevilla, Spain. Personal Communication, 2014.
- Devos, D.P. Regarding the presence of membrane coat proteins in bacteria: Confusion? What confusion? BioEssays 2012, 34, 38–39. [Google Scholar] [CrossRef] [PubMed]
- Budd, A.; Devos, D.P. Evaluating the evolutionary origins of unexpected character distributions within the bacterial Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Front. Evol. Genomic Microbiol. 2012, 3, 401. [Google Scholar]
- Lonhienne, T.G.A.; Sagulenko, E.; Webb, R.I.; Lee, K.-C.; Franke, J.; Devos, D.P.; Nouwens, A.; Carroll, B.J.; Fuerst, J.A. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc. Natl. Acad. Sci. USA 2010, 107, 12883–12888. [Google Scholar] [CrossRef] [PubMed]
- Kuper, U.; Meyer, C.; Muller, V.; Rachel, R.; Huber, H. Energized outer membrane and spatial separation of metabolic processes in the hyperthermophilic Archaeon Ignicoccus hospitalis. Proc. Natl. Acad. Sci. USA 2010, 107, 3152–3156. [Google Scholar] [CrossRef] [PubMed]
- Lane, N.; Martin, W. The energetics of genome complexity. Nature 2010, 467, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.A.; Foster, P.G.; Cox, C.J.; Embley, T.M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature 2013, 504, 231–236. [Google Scholar] [CrossRef] [PubMed]
- Kelly, S.; Wickstead, B.; Gull, K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the archaea and a thaumarchaeal origin for the eukaryotes. Proc. Biol. Sci. 2011, 278, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Mast, F.D.; Barlow, L.D.; Rachubinski, R.A.; Dacks, J.B. Evolutionary mechanisms for establishing eukaryotic cellular complexity. Trends Cell Biol. 2014, 24, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Schopf, J.W. Deep divisions in the tree of life—What does the fossil record reveal? Biol. Bull. 1999, 196, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Javaux, E.J. The early eukaryotic fossil record. Adv. Exp. Med. Biol. 2007, 607, 1–19. [Google Scholar] [PubMed]
- Javaux, E.J.; Marshall, C.P.; Bekker, A. Organic-walled microfossils in 3.2-billion-year-old shallow-marine siliciclastic deposits. Nature 2010, 463, 934–938. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C. The origin of eukaryotes: A reappraisal. Nat. Rev. Genet. 2007, 8, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Embley, T.M.; Martin, W. Eukaryotic evolution, changes and challenges. Nature 2006, 440, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, I.C. A phylum level perspective on bacterial cell envelope architecture. Trends Microbiol. 2010, 18, 464–470. [Google Scholar] [CrossRef] [PubMed]
- Neumann, N.; Lundin, D.; Poole, A.M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLOS ONE 2010, 5, e13241. [Google Scholar] [CrossRef] [PubMed]
- Gribaldo, S.; Poole, A.M.; Daubin, V.; Forterre, P.; Brochier-Armanet, C. The origin of eukaryotes and their relationship with the archaea: Are we at a phylogenomic impasse? Nat. Rev. Microbiol. 2010, 8, 743–752. [Google Scholar] [CrossRef] [PubMed]
- Devos, D.P.; Gräf, R.; Field, M.C. Evolution of the nucleus. Curr. Opin. Cell Biol. 2014, 28, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalier-Smith, T. Molecular phylogeny. Archaebacteria and archezoa. Nature 1989, 339, l00–101. [Google Scholar] [CrossRef] [PubMed]
- Poole, A.; Penny, D. Eukaryote evolution: Engulfed by speculation. Nature 2007, 447, 913. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle trafficking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Arechaga, I. Membrane invaginations in bacteria and mitochondria: Common features and evolutionary scenarios. J. Mol. Microbiol. Biotechnol. 2013, 23, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Cabré, E.J.; Sánchez-Gorostiaga, A.; Carrara, P.; Ropero, N.; Casanova, M.; Palacios, P.; Stano, P.; Jiménez, M.; Rivas, G.; Vicente, M. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J. Biol. Chem. 2013, 288, 26625–26634. [Google Scholar] [CrossRef] [PubMed]
- Diekmann, Y.; Pereira‑Leal, J.B. Evolution of intracellular compartmentalization. Biochem. J. 2013, 449, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Wessels, H.J.C.T.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Kartal, B.; Jetten, M.S.M.; van Niftrik, L. Isolation and characterization of a prokaryotic cell organelle from the anammox bacterium Kuenenia stuttgartiensis. Mol. Microbiol. 2014, 94, 794–802. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Sánchez, J.C.; Costa, R.; Devos, D.P. A Multi-Functional Tubulovesicular Network as the Ancestral Eukaryotic Endomembrane System. Biology 2015, 4, 264-281. https://doi.org/10.3390/biology4020264
González-Sánchez JC, Costa R, Devos DP. A Multi-Functional Tubulovesicular Network as the Ancestral Eukaryotic Endomembrane System. Biology. 2015; 4(2):264-281. https://doi.org/10.3390/biology4020264
Chicago/Turabian StyleGonzález-Sánchez, Juan Carlos, Ricardo Costa, and Damien P. Devos. 2015. "A Multi-Functional Tubulovesicular Network as the Ancestral Eukaryotic Endomembrane System" Biology 4, no. 2: 264-281. https://doi.org/10.3390/biology4020264
APA StyleGonzález-Sánchez, J. C., Costa, R., & Devos, D. P. (2015). A Multi-Functional Tubulovesicular Network as the Ancestral Eukaryotic Endomembrane System. Biology, 4(2), 264-281. https://doi.org/10.3390/biology4020264