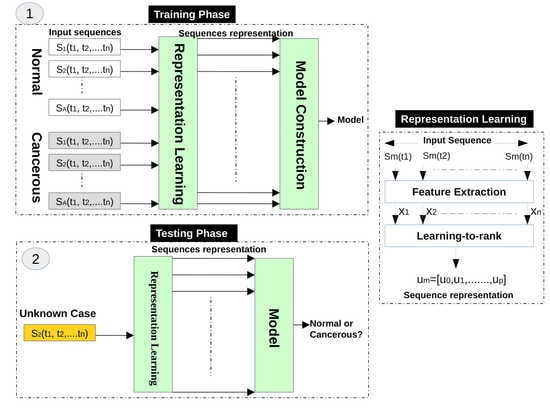
In this stage, we use six texture analysis methods to extract texture descriptors from each infrared image, and then the LTR method is exploited to generate a representation for the infrared images of each case.
3.1.1. Texture Analysis
To describe the texture of breast tissues in infrared images accurately, we assess the efficacy of six texture analysis methods: histogram of oriented gradients, lacunarity analysis of vascular networks, local binary pattern, local directional number pattern, Gabor filters and descriptors computed from the gray level co-occurrence matrix. These texture analysis methods have been widely used in medical image analysis and achieved good results with breast cancer image analysis [
16,
27,
28,
36,
37,
38].
Histogram of oriented gradients (HOG): The HOG method is considered as one of the most powerful texture analysis methods because it produces distinctive descriptors in the case of illumination changes and cluttered background [
39]. The HOG method has been used in several studies to analyze medical images. For instance, in [
16], a nipple detection algorithm was used to determine the ROIs and then the HOG method was used to detect breast cancer in infrared images. To construct the HOG descriptor, the occurrences of edge orientations in a local image neighborhood are accumulated. The input image is split into a set of blocks (each block includes small groups of cells). For each block, a weighted histogram is constructed, and then the frequencies in the histograms are normalized to compensate for the changes in illumination. Finally, the histograms of all blocks are concatenated to build the final HOG descriptor.
Lacunarity analysis of vascular networks (LVN): Lacunarity refers to a measure of how patterns clog the space [
40] and describes spatial features, multi-fractal and even non-fractal patterns. We use the lacunarity of vascular networks (LVN) in thermograms to detect breast cancer. To calculate LVN features, we first extract the vascular network (
) from each infrared image and then calculate the lacunarity-related features
. Please Please note thatLVN extracts 2 features from each infrared image. Below, we briefly explain the two stages:
- -
Vascular network extraction: this method has two steps [
27]:
This process produces a binary image (i.e., VN is a binary image). In
Figure 3, we show an example of a vascular network extracted from an infrared image.
- -
Lacunarity-related features: We use the sliding box method to determine the lacunarity on the binary image
. In this method, an
patch is moved through the image VN, and then we count the number of mass points within the patch at each position and compute a histogram
;
n denotes the number of mass points in each patch. The histogram
refers to the number of patches containing
n mass points. The lacunarity at scale
l for the pixel at the position
can be determined as follows [
40]:
where
calculates the expectation of the random variable
X. The study of [
41] demonstrated that the lacunarity shows power law behaviors with its scale
l as follows:
We take the logarithm on left and right sides of Equation (
2) to get the following equation:
To estimate
and
, we use the linear least squares fitting technique, and set the scale range to compute the lacunarity in the interval [
42,
43].
Gabor filters (GF): GF have been used in several works on medical image analysis. For instance, the authors of [
44] extracted Gabor features from breast thermograms, such as energy and amplitude in different scales and orientations to quantify the asymmetry between normal and abnormal breast tissues. In [
45], self-similar GF, the Karhunen–Loève (KL) transform and Otsu’s thresholding method were used to detect global signs of asymmetry in the fibro-glandular discs of the left and right mammograms of breast cancer patients. The KL transform was used to preserve the most relevant directional elements appearing at all scales. In [
37], GF were used to extract directional features from mammographic images and the principal component analysis method was used for dimensionality reduction. In [
46], GF were used to extract texture features from mammographic images to detect breast cancer.
The 2D Gabor filter
can be formulated as a sinusoidal with a certain frequency and orientation multiplied by a Gaussian envelope as follows:
where
,
and
are the standard deviations along the x- and y-axes, and the centre of the sinusoidal function, respectively. In our experiments, 4 scales and 6 orientations are used to compute GF responses (the number of scales and orientation were empirically tuned). Then, we convolve each input image
with each response
to determine the filtered image
. To extract texture features from GF, the energy is computed from each filtered image, and then all energies are concatenated into one feature vector [
47].
Local binary pattern (LBP): The LBP operator assigns a label (0 or 1) for each pixel in the input image. LBP extracts a
box around each pixel
and then compares all pixels with it. Pixels in this box with a value greater than
are labelled by ‘1’ and the other pixels are labelled by ‘0’. In this way, LBP represents each pixel with 8-bits [
36,
48]. We only consider patterns that comprise at most two transitions
from 1 to 0 or
from 0 to 1 (called uniform LBPs). Please Please note that
10101010 is a non-uniform LBP and
00111110 is a uniform LBP. To construct the final LBP descriptor, we compute the histogram of uniform patterns (its length is 59).
Local directional number (LDN): Rámirez et al. proposed the LDN descriptor in [
49], in which 8 Kirsch compass masks are convolved with the input image, yielding 8 edge responses for each pixel (each response is a result for a mask). The authors used a code for each mask (called location code): 000 for mask0, 001 for mask1, 010 for mask2, 011 for mask3, 100 for mask4, 101 for mask5, 110 for mask6 and 111 for mask7. For each pixel in the image, LDN determines the maximum positive and smallest negative edge responses, and then the location codes corresponding to the two responses are concatenated.
Assume that we convolve the eight masks (mask0–mask7) with an infrared image and we get eight edge responses (10, −20, 0, 3, 7, −10, 6, and 30) for the pixel . The maximum positive and smallest negative edge responses are 30 and −20 that correspond to the masks mask1 and mask7. Thus, the LDN code for the pixel is 111001. A 64-bin histogram is computed from the codes extracted from the input infrared images.
Features computed from the gray level co-occurrence matrix (GLCM): To compute the GLCM, we determine the joint frequencies
of possible combinations of gray levels
i and
j in the input image. Please Please note thateach pair is separated by a distance
and an angle
[
50]. In this research, we set the distance to 5 and use 4 different orientations (
,
,
and
) with a quantification level of 32 to compute 22 descriptors from each GLCM. This configuration was recommended in several related studies, such as [
38,
51]. We then concatenate all descriptors into one feature vector (its dimension is 88,
). In
Table 1, we present the descriptors extracted from the GLCM in addition to the mathematical expression of each descriptor [
50,
52,
53]. The interpretation of terms used to compute the descriptors are provided in
Table 2.
3.1.2. Representation Learning Using LTR
Based on the observation that the changes of temperatures on normal and abnormal tissues have a different temporal ordering, we use the LTR method to model the evolution of temperatures of women breasts during dynamic thermography procedures. In other words, we use the LTR method to generate a compact description for each image sequence. Assume that we have a sequence of thermograms of a breast (temperature matrices), and each thermogram at time
t is represented by a vector
, thus the whole sequence is represented by
; Nsq is the number of images in the sequence (in our case Nsq = 20). The feature vector of each thermogram
is obtained in the feature extraction stage (
Section 3.1.1).
We use the LTR method to model the relative rank of these thermograms (e.g., comes after , which can be written as ). Assume m3 is the feature vector of an infrared image acquired at time step t3, m2 is the feature vector of an infrared image acquired at time step t2, and m1 is the feature vector of an infrared image acquired at time step t1, this means that m3 comes after m2, and m2 comes after m1. We use the LTR method to model this ranking, which represents the change of temperature in breasts from a time step to another during the dynamic thermography procedure.
To avoid the effect of abrupt changes of temperatures in thermograms, the LTR method learns the order of smoothed versions of the feature vectors. Let us define a sequence
, where
is the result of processing the feature vectors from time 1 to
t (
to
) using the time varying mean method as follows:
Each vector
is then normalized as follows:
where
is the normalized version of
. Given the smoothed vectors, LTR learns their rank (i.e.,
), a linear LTR learns a pairwise linear function
, where
is a vector containing its parameters. The ranking score of
is computed by
. The LTR method optimizes the parameters
of
using the objective function of Equation (
7) with the constraint
as follows [
54]:
where
is the regularization parameter and
is the margin of tolerance. The constraint of the objective function encourages the ranking score (prediction score) of the difference between the feature vectors of two infrared images
and
acquired at time
and
, respectively, to be greater than
, and the margin of tolerance should be greater than or equal zero. In other words, if
comes after
, the ranking score of
should be greater than the one of
. For further details the reader is referred to [
55,
56]. In our case, the temperature evolution information is encoded in the parameters
, which can be viewed as a principled, data-driven, temporal pooling of the evolution of temperature in thermography procedures. Thus, the parameters
are used in this study to describe the evolution of temperatures in the whole sequence of thermograms. In other words, we use
to represent the whole sequence. In this study, we model the evolution of temperatures in thermography procedures in the forward and backward directions, as follows:
Forward direction (): we model the changes in the temperature from the cooling instant to the instant of restoring the normal temperature of the body (the algorithm moves from the infrared image acquired at time step t to the image acquired at ).
Backward direction (): we model the changes in temperature in the reverse direction where we start with the last infrared image of the input sequence and proceed to the first one (the algorithm moves from the infrared image acquired at time step t to the image acquired at ).
In our experiments, we calculate the parameters of LTR two times for each input sequence: one for the forward direction and another for the backward direction . Prior to the classification stage, we normalize each feature vector using the norm. In the classification stage, we input the feature vectors of normal and cancerous cases into the MLP classifier to construct a classification model.