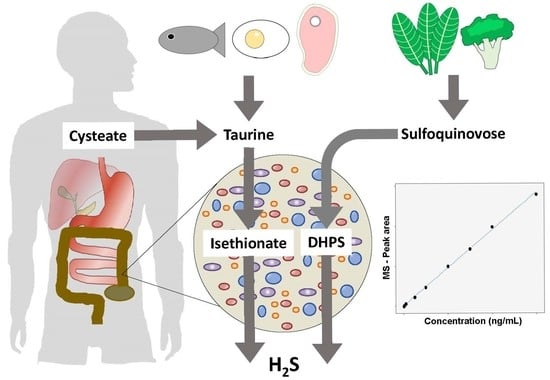
Multiplexed Quantitative Assessment of the Fate of Taurine and Sulfoquinovose in the Intestinal Microbiome
Abstract
:1. Introduction
2. Results
2.1. Method Optimization and Validation
2.2. Application: The Fate of SQ in the SIHUMI Consortium with and without Bilophila wadsworthia
2.3. Application: The SIHUMI Consortium Is Only Able to Degrade Taurine to Sulfide in the Presence of Bilophila wadsworthia
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Collection and Processing of Fecal Samples
4.3. Replicates
4.4. LC-MS/MS-MRM Method
4.5. Method Validation
4.6. Bacterial Growth Conditions
4.7. Bacterial Incubation Experiments
4.8. Bacterial Sample Preparation for Sulfonate Quantification
4.9. Sulfide Quantification
4.10. Statistics for Application Experiment
4.11. LC-MS/MS-MRM Data Availability
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Heintz-Buschart, A.; Wilmes, P. Human Gut Microbiome: Function Matters. Trends Microbiol. 2018, 26, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carbonero, F.; Benefiel, A.; Alizadeh-Ghamsari, A.; Gaskins, H.R. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front. Physiol. 2012, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.B.; Lin, H.C. Hydrogen Sulfide in Physiology and Diseases of the Digestive Tract. Microorganisms 2015, 3, 866–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Speciale, G.; Jin, Y.; Davies, G.J.; Williams, S.J.; Goddard-Borger, E.D. YihQ is a sulfoquinovosidase that cleaves sulfoquinovosyl diacylglyceride sulfolipids. Nat. Chem. Biol. 2016, 12, 215–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denger, K.; Weiss, M.; Felux, A.K.; Schneider, A.; Mayer, C.; Spiteller, D.; Huhn, T.; Cook, A.M.; Schleheck, D. Sulphoglycolysis in Escherichia coli K-12 closes a gap in the biogeochemical sulphur cycle. Nature 2014, 507, 114–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard-Borger, E.D.; Williams, S.J. Sulfoquinovose in the biosphere: Occurrence, metabolism and functions. Biochem. J. 2017, 474, 827–849. [Google Scholar] [CrossRef] [PubMed]
- Sacoman, J.L.; Badish, L.N.; Sharkey, T.D.; Hollingsworth, R.I. The metabolic and biochemical impact of glucose 6-sulfonate (sulfoquinovose), a dietary sugar, on carbohydrate metabolism. Carbohydr. Res. 2012, 362, 21–29. [Google Scholar] [CrossRef]
- Laidlaw, S.; Grosvenor, M.; Kopple, J. The taurine content of common foodstuffs. J. Parenter. Enter. Nutr. 1990, 14, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Dawson, P.A.; Karpen, S.J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 2015, 56, 1085–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahlström, A.; Sayin, S.I.; Marschall, H.-U.; Bäckhed, F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016, 24, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.W.; Qiao, S.Y.; Li, D.F. Amino acids and gut function. Amino acids 2009, 37, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ai, Y.; Niu, X.; Shang, F.; Li, Z.; Liu, H.; Li, W.; Ma, W.; Chen, R.; Wei, T.; et al. Taurine protects against cardiac dysfunction induced by pressure overload through SIRT1–p53 activation. Chem. Biol. Interact. 2020, 317, 108972. [Google Scholar] [CrossRef]
- Lambert, I.H.; Kristensen, D.M.; Holm, J.B.; Mortensen, O.H. Physiological role of taurine–From organism to organelle. Acta Physiol. 2015, 213, 191–212. [Google Scholar] [CrossRef] [PubMed]
- Qaradakhi, T.; Gadanec, L.K.; McSweeney, K.R.; Abraham, J.R.; Apostolopoulos, V.; Zulli, A. The Anti-Inflammatory Effect of Taurine on Cardiovascular Disease. Nutrients 2020, 12, 2847. [Google Scholar] [CrossRef]
- Felux, A.-K.; Spiteller, D.; Klebensberger, J.; Schleheck, D. Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1. Proc. Natl. Acad. Sci. USA 2015, 112, E4298–E4305. [Google Scholar] [CrossRef] [Green Version]
- Barton, L.L.; Ritz, N.L.; Fauque, G.D.; Lin, H.C. Sulfur Cycling and the Intestinal Microbiome. Dig. Dis. Sci. 2017, 62, 2241–2257. [Google Scholar] [CrossRef]
- Gallego, D.; Clavé, P.; Donovan, J.; Rahmati, R.; Grundy, D.; Jiménez, M.; Beyak, M.J. The gaseous mediator, hydrogen sulphide, inhibits in vitro motor patterns in the human, rat and mouse colon and jejunum. Neurogastroenterol. Motil. 2008, 20, 1306–1316. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Liang, F.; Shah Masood, W.; Yan, X. Hydrogen sulfide protected gastric epithelial cell from ischemia/reperfusion injury by Keap1 s-sulfhydration, MAPK dependent anti-apoptosis and NF-κB dependent anti-inflammation pathway. Eur. J. Pharmacol. 2014, 725, 70–78. [Google Scholar] [CrossRef]
- Mok, Y.Y.; Moore, P.K. Hydrogen sulphide is pro-inflammatory in haemorrhagic shock. Inflamm. Res. 2008, 57, 512–518. [Google Scholar] [CrossRef]
- Tomasova, L.; Konopelski, P.; Ufnal, M. Gut Bacteria and Hydrogen Sulfide: The New Old Players in Circulatory System Homeostasis. Molecules 2016, 21, 1558. [Google Scholar] [CrossRef] [PubMed]
- Peck, S.C.; Denger, K.; Burrichter, A.; Irwin, S.M.; Balskus, E.P.; Schleheck, D. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc. Natl. Acad. Sci. USA 2019, 116, 3171–3176. [Google Scholar] [CrossRef] [Green Version]
- Attene-Ramos, M.S.; Nava, G.M.; Muellner, M.G.; Wagner, E.D.; Plewa, M.J.; Gaskins, H.R. DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 Int cells. Environ. Mol. Mutagen. 2010, 51, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Ijssennagger, N.; van der Meer, R.; van Mil, S.W.C. Sulfide as a Mucus Barrier-Breaker in Inflammatory Bowel Disease? Trends Mol. Med. 2016, 22, 190–199. [Google Scholar] [CrossRef]
- Agnello, G.; Chang, L.L.; Lamb, C.M.; Georgiou, G.; Stone, E.M. Discovery of a substrate selectivity motif in amino acid decarboxylases unveils a taurine biosynthesis pathway in prokaryotes. ACS Chem. Biol. 2013, 8, 2264–2271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, M.; Wei, Y.; Zhou, Y.; Zhang, J.; Lin, L.; Hu, Y.; Hua, G.; N Nanjaraj Urs, A.; Liu, D.; Wang, F.; et al. Radical-mediated C-S bond cleavage in C2 sulfonate degradation by anaerobic bacteria. Nat. Commun. 2019, 10, 1609. [Google Scholar] [CrossRef]
- Burrichter, A.; Denger, K.; Franchini, P.; Huhn, T.; Müller, N.; Spiteller, D.; Schleheck, D. Anaerobic Degradation of the Plant Sugar Sulfoquinovose Concomitant With H(2)S Production: Escherichia coli K-12 and Desulfovibrio sp. Strain DF1 as Co-culture Model. Front. Microbiol. 2018, 9, 2792. [Google Scholar] [CrossRef] [Green Version]
- Becker, N.; Kunath, J.; Loh, G.; Blaut, M. Human intestinal microbiota: Characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2011, 2, 25–33. [Google Scholar] [CrossRef]
- Denger, K.; Laue, H.; Cook, A.M. Anaerobic taurine oxidation: A novel reaction by a nitrate-reducing Alcaligenes sp. Microbiology 1997, 143, 1919–1924. [Google Scholar] [CrossRef] [Green Version]
- Laue, H.; Denger, K.; Cook, A.M. Fermentation of cysteate by a sulfate-reducing bacterium. Arch. Microbiol. 1997, 168, 210–214. [Google Scholar] [CrossRef] [Green Version]
- Styp von Rekowski, K.; Denger, K.; Cook, A.M. Isethionate as a product from taurine during nitrogen-limited growth of Klebsiella oxytoca TauN1. Arch. Microbiol. 2005, 183, 325–330. [Google Scholar] [CrossRef] [Green Version]
- Siskos, A.P.; Jain, P.; Römisch-Margl, W.; Bennett, M.; Achaintre, D.; Asad, Y.; Marney, L.; Richardson, L.; Koulman, A.; Griffin, J.L.; et al. Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma. Anal. Chem. 2017, 89, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Karp, P.D.; Billington, R.; Caspi, R.; Fulcher, C.A.; Latendresse, M.; Kothari, A.; Keseler, I.M.; Krummenacker, M.; Midford, P.E.; Ong, Q.; et al. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 2017, 20, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- van der Ploeg, J.R.; Eichhorn, E.; Leisinger, T. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch. Microbiol. 2001, 176, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, E.; van der Ploeg, J.R.; Kertesz, M.A.; Leisinger, T. Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 1997, 272, 23031–23036. [Google Scholar] [CrossRef] [Green Version]
- da Silva, S.M.; Venceslau, S.S.; Fernandes, C.L.; Valente, F.M.; Pereira, I.A. Hydrogen as an energy source for the human pathogen Bilophila wadsworthia. Antonie Van Leeuwenhoek 2008, 93, 381–390. [Google Scholar] [CrossRef]
- Moench, T.T.; Zeikus, J.G. An improved preparation method for a titanium (III) media reductant. J. Microbiol. Methods 1983, 1, 199–202. [Google Scholar] [CrossRef]
- Strocchi, A.; Furne, J.K.; Levitt, M.D. A modification of the methylene blue method to measure bacterial sulfide production in feces. J. Microbiol. Methods 1992, 15, 75–82. [Google Scholar] [CrossRef]
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]





| Sulfonate | Q1 Mass (Da) | Q3 Mass (Da) | Time (ms) | ID | DP (V) | CE (V) |
|---|---|---|---|---|---|---|
| Cysteate | 168.2 | 151 | 20 | Cysteate_1 * | −80 | −17 |
| 168.2 | 86 | 20 | Cysteate_2 | −80 | −18 | |
| 168.2 | 81 | 20 | Cysteate_3 | −80 | −27 | |
| 168.2 | 71 | 20 | Cysteate_4 | −80 | −24 | |
| Isethionate | 125.3 | 107 | 20 | Ise_1 | −90 | −21 |
| 125.3 | 95 | 20 | Ise_2 * | −90 | −20 | |
| 125.3 | 80 | 20 | Ise_3 | −90 | −32 | |
| 2,3-dihydroxy-1-propanesulfonate | 155 | 95 | 20 | DHPS_1 * | −96 | −24 |
| 155 | 80 | 20 | DHPS_2 | −96 | −39 | |
| Sulfoquinovose | 243 | 123 | 20 | SQ_1 * | −110 | −31 |
| 243 | 95 | 20 | SQ_2 | −110 | −44 | |
| 243 | 153 | 20 | SQ_3 | −110 | −24 | |
| 243 | 183 | 20 | SQ_4 | −110 | −23 | |
| 243 | 81 | 20 | SQ_5 | −110 | −32 | |
| Taurine | 124 | 81 | 20 | Taurine_1 * | −95 | −30 |
| 124 | 80 | 20 | Taurine_2 | −100 | −20 | |
| 124 | 65 | 20 | Taurine_3 | −100 | −19 |
| In Extracted Diluted Measured Sample | In Fecal Supernatant | |||||
|---|---|---|---|---|---|---|
| Sulfonate | LLOD (µM) | LLOQ (µM) | ULOQ (µM) | Molar Mass (g/mol) | LLOQ (mM) | ULOQ (mM) |
| Cysteate | 0.0015 | 0.003 | 3.55 | 169.15 | 0.03 | 35.5 |
| Isethionate | 0.0079 | 0.024 | 4.76 | 126.13 | 0.24 | 47.6 |
| 2,3-dihydroxy-1-propanesulfonate | 0.0006 | 0.0016 | 3.87 | 155.15 | 0.016 | 38.7 |
| Sulfoquinovose | 0.001 | 0.002 | 2.46 | 244.22 | 0.02 | 24.6 |
| Taurine | 0.001 | 0.002 | 4.8 | 125.14 | 0.02 | 48.0 |
| Sulfonate | Theoretical Concentration (µM) | Intraday Mean Recovery (%) | Intraday RSD (%) | Interday Mean Recovery (%) | Interday RSD (%) | N |
|---|---|---|---|---|---|---|
| 2,3-dihydroxy-1-propanesulfonate | 0.0016 | 95.2 | 8.2 | 106.2 | 8.1 | 7 |
| 0.0048 | 94..4 | 9.4 | 104.4 | 8.7 | 7 | |
| 0.0097 | 94.4 | 9.3 | 106.9 | 5.1 | 7 | |
| 1.933 | 96.7 | 6.5 | 107.2 | 11.2 | 7 | |
| 2.9 | 96 | 12.1 | 111.1 | 12.9 | 7 | |
| Taurine | 0.002 | 102.9 | 5.5 | 111.6 | 5.5 | 7 |
| 0.006 | 104.4 | 6.5 | 113.6 | 10.7 | 7 | |
| 0.012 | 103.4 | 6,3 | 116.6 | 10.4 | 7 | |
| 2.4 | 95.9 | 2.4 | 106.7 | 9.3 | 7 | |
| 3.6 | 91.2 | 9.9 | 115 | 28 | 7 | |
| Cysteate | 0.003 | 87 | 15.7 | 95.8 | 9.9 | 7 |
| 0.009 | 89.1 | 11.1 | 98..4 | 4.6 | 7 | |
| 0.0177 | 95.5 | 6.9 | 98.5 | 11.4 | 7 | |
| 1.773 | 97.1 | 4.8 | 106.7 | 8.3 | 7 | |
| 2.66 | 94.8 | 8.2 | 103.9 | 5 | 7 | |
| Isethionate | 0.024 | 73.3 | 9 | 129.3 | 20.3 | 7 |
| 0.079 | 79.1 | 8.8 | 133.2 | 22.1 | 7 | |
| 0.396 | 81.4 | 4.8 | 128.1 | 22.6 | 7 | |
| 2.37 | 85.5 | 7.2 | 117.8 | 18.8 | 7 | |
| 3.57 | 79.7 | 9.1 | 114.3 | 15.5 | 7 | |
| Sulfoquinovose | 0.002 | 94.8 | 20 | 109.4 | 15.5 | 7 |
| 0.006 | 94.8 | 7.1 | 103.3 | 11.9 | 7 | |
| 0.012 | 92.3 | 7.6 | 104.1 | 15.5 | 7 | |
| 1.228 | 98.1 | 3.4 | 114.1 | 15.9 | 7 | |
| 1.843 | 96.6 | 6.4 | 112 | 14.3 | 7 |
| Solution | Components with Concentration |
|---|---|
| Anoxic phosphate-buffered saline (PBS) | 8.5 g/L NaCl |
| 0.3 g/L KH2PO4 | |
| 0.6 g/L Na2HPO4 | |
| 0.1 g/L bacteriological peptone | |
| 1 mg/L resazurin | |
| 40 mM sodium DL-lactate | |
| 40 mM sodium formate | |
| pH 7.0 | |
| N2/CO2 (80/20, v/v) as gas phase, | |
| autoclaved at 121 °C for 15 min | |
| Trace-element solution | 10 mL/L HCl |
| 1.5 g/L FeCl2 × 4 H2O | |
| 70 mg/L ZnCl2 | |
| 100 mg/L MnCl2 × 4 H2O | |
| 6 mg/L H3BO3 | |
| 190 mg/L CoCl2 × 6 H2O | |
| 2 mg/L CuCl2 × 2 H2O | |
| 24 mg/L NiCl2 × 6 H2O | |
| 36 mg/L Na2MoO4 × 2 H2O | |
| Selenite–tungstate solution | 500 mg/L NaOH |
| 3 mg/L Na2SeO3 × 5 H2O | |
| 4 mg/L Na2WO4 × 2 H2O | |
| Seven-vitamin solution | 100 mg/L Vitamin B12 |
| 80 mg/L p-amino benzoic acid | |
| 20 mg/L D (+)-biotin | |
| 200 mg/L nicotinic acid | |
| 100 mg/L calcium pantothenate | |
| 300 mg/L pyridoxine hydrochloride | |
| 200 mg/L thiamine hydrochloride × 2 H2O | |
| Ti(III) nitrilotriacetate solution | 19.2 g/L nitrilotriacetic acid diluted in anoxic distilled water, |
| pH of 9 adjusted with NaOH | |
| 19.2 mL 20% TiCl3 (Acros), | |
| pH of 7 adjusted with Na2CO3 (80 g/L) |
| Bacterial Species | Strain Designation (T Type Strain) | Cell Number in 10 mL DS Medium |
|---|---|---|
| Anaerostipes caccae | DSM 14662T | 107 |
| Bifidobacterium longum | NCC 2705 | 106 |
| Blautia producta | DSM 2950T | 109 |
| Bacteroides thetaiotaomicron | DSM 2079T | 1010 |
| Clostridium butyricum | DSM 10702T | 107 |
| Clostridium ramosum | DSM 1402T | 109 |
| Escherichia coli K-12 | MG1655 | 109 |
| Lactobacillus plantarum | DSM 20174T | 103 |
| Bilophila wadsworthia | DSM 11045 | 109 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haange, S.-B.; Groeger, N.; Froment, J.; Rausch, T.; Burkhardt, W.; Gonnermann, S.; Braune, A.; Blaut, M.; von Bergen, M.; Rolle-Kampczyk, U. Multiplexed Quantitative Assessment of the Fate of Taurine and Sulfoquinovose in the Intestinal Microbiome. Metabolites 2020, 10, 430. https://doi.org/10.3390/metabo10110430
Haange S-B, Groeger N, Froment J, Rausch T, Burkhardt W, Gonnermann S, Braune A, Blaut M, von Bergen M, Rolle-Kampczyk U. Multiplexed Quantitative Assessment of the Fate of Taurine and Sulfoquinovose in the Intestinal Microbiome. Metabolites. 2020; 10(11):430. https://doi.org/10.3390/metabo10110430
Chicago/Turabian StyleHaange, Sven-Bastiaan, Nicole Groeger, Jean Froment, Theresa Rausch, Wiebke Burkhardt, Svenja Gonnermann, Annett Braune, Michael Blaut, Martin von Bergen, and Ulrike Rolle-Kampczyk. 2020. "Multiplexed Quantitative Assessment of the Fate of Taurine and Sulfoquinovose in the Intestinal Microbiome" Metabolites 10, no. 11: 430. https://doi.org/10.3390/metabo10110430
APA StyleHaange, S. -B., Groeger, N., Froment, J., Rausch, T., Burkhardt, W., Gonnermann, S., Braune, A., Blaut, M., von Bergen, M., & Rolle-Kampczyk, U. (2020). Multiplexed Quantitative Assessment of the Fate of Taurine and Sulfoquinovose in the Intestinal Microbiome. Metabolites, 10(11), 430. https://doi.org/10.3390/metabo10110430