Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum lycopersicum Plants Responding to Phytophthora capsici
Abstract
:1. Introduction
2. Results
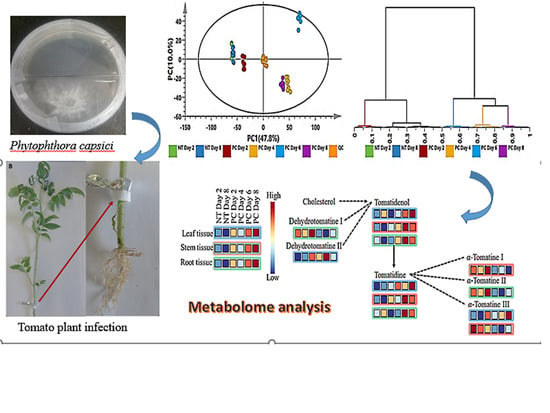
2.1. Symptom Development
2.2. Aromatic Amino Acid Quantification
2.3. Quantification of Methyl Salicylic Acid and 1-Aminocyclopropane-1-Carboxylic Acid
2.4. Metabolic Profiling of P. capsici-Induced Changes in Tomato Plants
2.4.1. Multivariate Data Analysis
2.4.2. Correlation Analysis of OPLS-DA-Derived Features from Control and Infected Tomato Plants
2.4.3. Time-Course of Comparative Metabolite Reprogramming in P. capsici-Infected Tomato Plants
Differential Reprogramming in Primary Metabolism
Differential Reprogramming of Phytohormones and Signalling Molecules
Differential Reprogramming of Flavonoids and Hydroxycinnamic Acid Derivatives
Differential Reprogramming of Glycoalkaloids
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions and Treatment with P. capsici
4.2. Targeted Metabolomics Analysis
4.3. Non-Targeted Metabolomics Analysis
4.3.1. Metabolite Extraction and Data Acquisition on an UHPLC-ESI-qTOF-MS (Ultra High-Performance Liquid Chromatography, Electrospray Ionisation, Quadrupole Time-of-Flight Mass Spectrometry)
4.3.2. Multivariate Data Analysis
4.3.3. Metabolite Annotation
4.3.4. Metabolite- Metabolite Correlation and Time-Dependent Reprogramming
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Hardham, A.R. The cell biology behind Phytophthora pathogenicity. Aust. Plant Pathol. 2001, 30, 91–98. [Google Scholar] [CrossRef]
- Fawke, S.; Doumane, M.; Schornack, S. Oomycete interactions with plants: Infection strategies and resistance principles. Microbiol. Mol. Biol. Rev. 2015, 79, 263–280. [Google Scholar] [CrossRef] [Green Version]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrath, P.G.U.; Beckers, G.J.M.; Flors, V.; García-Agustín, P.; Jakab, G.; Mauch, F.; Newman, M.; Pieterse, C.M.J.; Poinssot, B.; Pozo, M.J.; et al. Priming: Getting ready for battle. Mol. Plant Microbe Interact. 2009, 19, 1062–1071. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol. Plant Pathol. 2011, 12, 702–708. [Google Scholar] [CrossRef]
- Liu, W.; Liu, J.; Ning, Y.; Ding, B.; Wang, X.; Wang, Z.; Wang, G.L. Recent progress in understanding PAMP- and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Mol. Plant 2013, 6, 605–620. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.A.; Sundelin, T.; Nielsen, J.T.; Erbs, G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front. Plant Sci. 2013, 4, 139. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Rapid and sensitive hormonal profiling of complex plant samples by liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Plant Methods 2011, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Suarez-Rodriguez, M.C.; Adams-Phillips, L.; Liu, Y.; Wang, H.; Su, S.H.; Jester, P.J.; Zhang, S.; Bent, A.F.; Krysan, P.J. MEKK1 is required for flg22-induced MPK4 activation in Arabidopsis plants. Plant Physiol. 2007, 143, 661–669. [Google Scholar] [CrossRef] [Green Version]
- Dempsey, D.A.; Klessig, D.F. SOS—Too many signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Zhang, L.; Liu, L.; Wang, J.; Li, C.; Wang, Q. Multiple phytohormone signalling pathways modulate susceptibility of tomato plants to Alternaria alternata f. sp. lycopersici. J. Exp. Bot. 2013, 64, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dempsey, D.A.; Klessig, D.F. How does the multifaceted plant hormone salicylic acid combat disease in plants and are similar mechanisms utilized in humans? BMC Biol. 2017, 15, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van Der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tugizimana, F.; Steenkamp, P.A.; Piater, L.A.; Dubery, I.A. Multi-platform metabolomic analyses of ergosterol-induced dynamic changes in Nicotiana tabacum cells. PLoS ONE 2014, 9, 87846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, D.; Flors, V.; Glauser, G.; Mauch-Mani, B. Metabolomics of cereals under biotic stress: Current knowledge and techniques. Front. Plant Sci. 2013, 4, 82. [Google Scholar] [CrossRef] [Green Version]
- Glazebrook, J. Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr. Opin. Plant Biol. 2001, 4, 301–308. [Google Scholar] [CrossRef]
- De Vos, M.; Van Oosten, V.R.; Van Poecke, R.M.P.; Van Pelt, J.A.; Pozo, M.J.; Mueller, M.J.; Buchala, A.J.; Métraux, J.; Van Loon, L.C.; Dicke, M.; et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol. Plant Microbe Interact. 2005, 18, 923–937. [Google Scholar] [CrossRef] [Green Version]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling1. Plant Physiol. 2008, 146, 839–844. [Google Scholar] [CrossRef] [Green Version]
- Verhage, A.; Van Wees, S.C.M.; Pieterse, C.M.J. Plant immunity: It’s the hormones talking, but what do they say? Plant Physiol. 2010, 154, 536–540. [Google Scholar] [CrossRef]
- Naseem, M.; Dandekar, T. The role of auxin-cytokinin antagonism in plant-pathogen interactions. PLoS Pathog. 2012, 8, 1003026. [Google Scholar] [CrossRef] [PubMed]
- Hardham, A.R.; Jones, D.A.; Takemoto, D. Cytoskeleton and cell wall function in penetration resistance. Curr. Opin. Plant Biol. 2007, 10, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Scheideler, M.; Schlaich, N.L.; Fellenberg, K.; Beissbarth, T.; Hauser, N.C.; Vingron, M.; Slusarenko, A.J.; Hoheisel, D. Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 2002, 277, 10555–10561. [Google Scholar] [CrossRef] [Green Version]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef]
- Bolton, M.D. Primary metabolism and plant defense—Fuel for the fire. Mol. Plant Microbe Interact. 2009, 22, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Ballester, A.R.; Lafuente, M.T.; De Vos, R.C.H.; Bovy, A.G.; González-Candelas, L. Citrus phenylpropanoids and defence against pathogens. Part I: Metabolic profiling in elicited fruits. Food Chem. 2013, 136, 178–185. [Google Scholar] [CrossRef] [Green Version]
- Widhalm, J.R.; Dudareva, N. A familiar ring to it: Biosynthesis of plant benzoic acids. Mol. Plant 2015, 8, 83–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parthasarathy, A.; Cross, P.J.; Dobson, R.C.J.; Adams, L.E.; Savka, M.A.; Hudson, A.O. A three-ring circus: Metabolism of the three proteogenic aromatic amino acids and their role in the health of plants and animals. Front. Mol. Biosci. 2018, 5, 29. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. J. 2011, 3, 001438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Idris, E.E.; Iglesias, D.J.; Talon, M.; Borriss, R. Tryptophan-dependent production of Indole-3-Acetic Acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 2007, 20, 619–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kangasjärvi, S.; Neukermans, J.; Li, S.; Aro, E.M.; Noctor, G. Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 2012, 63, 1619–1636. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Piater, L.A.; Dubery, I.A. Plant metabolomics: A new frontier in phytochemical analysis. S. Afr. J. Sci. 2013, 109, 18–20. [Google Scholar] [CrossRef]
- Xu, Y.; Correa, E.; Goodacre, R. Integrating multiple analytical platforms and chemometrics for comprehensive metabolic profiling: Application to meat spoilage detection. Anal. Bioanal. Chem. 2013, 405, 5063–5074. [Google Scholar] [CrossRef]
- Quesada-Ocampo, L.M.; Vargas, A.M.; Naegele, R.P.; Francis, D.M.; Hausbeck, M.K. Resistance to crown and root rot caused by Phytophthora capsici in a Tomato advanced backcross of Solanum habrochaites and Solanum lycopersicum. Plant Dis. 2015, 100, 829–835. [Google Scholar] [CrossRef] [Green Version]
- Jupe, J.; Stam, R.; Howden, A.J.M.; Morris, J.A.; Zhang, R.; Hedley, P.E.; Huitema, E. Phytophthora capsici-tomato interaction features dramatic shifts in gene expression associated with a hemi-biotrophic lifestyle. Genome Biol. 2013, 14, R63. [Google Scholar] [CrossRef] [Green Version]
- Worley, B.; Halouska, S.; Powers, R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 2013, 433, 102–104. [Google Scholar] [CrossRef] [Green Version]
- Tugizimana, F.; Steenkamp, P.; Piater, L.; Dubery, I. A conversation on data mining strategies in LC-MS untargeted metabolomics: Pre-processing and pre-treatment steps. Metabolites 2016, 6, 40. [Google Scholar] [CrossRef] [Green Version]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar]
- Gabor, J.S. Hierarchical clustering via joint-within diastances: Extending ward’s minimum variance and method. J. Classif. 2005, 22, 151–183. [Google Scholar] [CrossRef]
- Fouedjio, F. A hierarchical clustering method for multivariate geostatistical data. Spat. Stat. 2016, 18, 333–351. [Google Scholar] [CrossRef]
- Granato, D.; Santos, J.S.; Escher, G.B.; Ferreira, B.L.; Maggio, R.M. Use of principal component analysis (PCA) and hierarchical cluster analysis (HCA) for multivariate association between bioactive compounds and functional properties in foods: A critical perspective. Trends Food Sci. Technol. 2018, 72, 83–90. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. A chemometrics toolbox based on projections and latent variables. J. Chemom. 2014, 28, 332–346. [Google Scholar] [CrossRef]
- Sumner, L.W.; Samuel, T.; Noble, R.; Gmbh, S.D.; Barrett, D.; Beale, M.H.; Hardy, N. Proposed minimum reporting standards for chemical analysis chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics 2013, 3, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Creek, D.J.; Dunn, W.B.; Fiehn, O.; Griffin, J.L.; Hall, R.D.; Lei, Z.; Mistrik, R.; Neumann, S.; Schymanski, E.L.; Sumner, L.W.; et al. Metabolite identification: Are you sure? And how do your peers gauge your confidence? Metabolomics 2014, 10, 350–353. [Google Scholar] [CrossRef]
- Boevink, P.C.; Birch, P.R.J.; Turnbull, D.; Whisson, S.C. Devastating intimacy: The cell biology of plant-Phytophthora interactions. New Phytol. 2020, 228, 445–458. [Google Scholar] [CrossRef]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef]
- Tzin, V.; Galili, G. The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. Arab. Book Am. Soc. Plant Biol. 2010, 8, 0132. [Google Scholar]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Priming agents of plant defence stimulate the accumulation of mono- and di-acylated quinic acids in cultured tobacco cells. Physiol. Mol. Plant Pathol. 2014, 88, 61–66. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Phenylpropanoid defences in Nicotiana tabacum cells: Overlapping metabolomes indicate common aspects to priming responses induced by lipopolysaccharides, chitosan and flagellin-22. PLoS ONE 2016, 11, 0151350. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant J. 2010, 63, 443–457. [Google Scholar] [CrossRef]
- Suharti, W.S.; Nose, A.; Zheng, S.H. Metabolite profiling of sheath blight disease resistance in rice: In the case of positive ion mode analysis by CE/TOF-MS. Plant Prod. Sci. 2016, 19, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Camañes, G.; Scalschi, L.; Vicedo, B.; González-Bosch, C.; García-Agustín, P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 2015, 84, 125–139. [Google Scholar]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing enviromental. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef] [Green Version]
- Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant. Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef]
- Ishihara, A.; Hashimoto, Y.; Tanaka, C.; Dubouzet, J.G.; Nakao, T.; Matsuda, F.; Nishioka, T.; Miyagawa, H.; Wakasa, K. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008, 54, 481–495. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.J.; Allwood, J.W.; Marsden-Edwards, E.; Putten, W.H.; Goodacre, R.; Dam, N.M. Metabolomic analysis of the interaction between plants and herbivores. Metabolomics 2008, 5, 150–161. [Google Scholar] [CrossRef] [Green Version]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Metabolomic profiling of the host response of tomato (Solanum lycopersicum) following infection by Ralstonia solanacearum. Int. J. Mol. Sci. 2019, 20, 3945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darandeh, N.; Hadavi, E. Effect of pre-harvest foliar application of citric acid and malic acid on chlorophyll content and post-harvest vase life of Lilium cv. Brunello. Front. Plant Sci. 2012, 2, 2011–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balmer, A.; Pastor, V.; Glauser, G.; Mauch-Mani, B. Tricarboxylates induce defense priming against bacteria in Arabidopsis thaliana. Front. Plant Sci. 2018, 9, 1–15. [Google Scholar]
- Fernie, A.R.; Carrari, F.; Sweetlove, L.J. Respiratory metabolism: Glycolysis, the TCA cycle and mitochondrial electron transport. Curr. Opin. Plant Biol. 2004, 7, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Garaude, J. Reprogramming of mitochondrial metabolism by innate immunity. Curr. Opin. Immunol. 2019, 56, 17–23. [Google Scholar] [CrossRef]
- Zeiss, D.R.; Mhlongo, M.I.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A. Comparative metabolic phenotyping of tomato (Solanum lycopersicum) for the identification of metabolic signatures in cultivars differing in resistance to Ralstonia solanacearum. Int. J. Mol. Sci. 2018, 19, 2558. [Google Scholar] [CrossRef] [Green Version]
- Kohli, A.; Sreenivasulu, N.; Lakshmanan, P.; Kumar, P.P. The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 2013, 32, 945–957. [Google Scholar] [CrossRef]
- Shah, J.; Chaturvedi, R.; Chowdhury, Z.; Venables, B.; Petros, R. Signaling by small metabolites in systemic acquired resistance. Plant J. 2014, 74, 645–658. [Google Scholar] [CrossRef]
- Manosalva, P.M.; Park, S.W.; Forouhar, F.; Tong, L.; Fry, W.E.; Klessig, D.F. Methyl esterase 1 (StMES1) is required for systemic acquired resistance in potato. Mol. Plant Microbe Interact. 2010, 23, 1151–1163. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lund, S.T.; Stall, R.E.; Klee, H.J. Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 1998, 10, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Medina, A.; Fernández, I.; Sánchez-Guzmán, M.J.; Jung, S.C.; Pascual, J.A.; Pozo, M.J. Deciphering the hormonal signalling network behind the systemic resistance induced by Trichoderma harzianum in tomato. Front. Plant Sci. 2013, 4, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riet, K.B.; Ndlovu, N.; Piater, L.A.; Dubery, I.A. Simultaneous analysis of defense-related phytohormones in Arabidopsis thaliana responding to fungal infection. Appl. Plant Sci. 2016, 4, 1600013. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.X.; Rossi, M.J.; Glick, B.R. Ethylene and 1-aminocyclopropane-1-carboxylate (ACC) in plant–bacterial interactions. Front. Plant Sci. 2018, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Ravanbakhsh, M.; Sasidharan, R.; Voesenek, L.A.C.J.; Kowalchuk, G.A.; Jousset, A. Microbial modulation of plant ethylene signaling: Ecological and evolutionary consequences. Microbiome 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Vanderstraeten, L.; Van Der Straeten, D. Accumulation and transport of 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: Current status, considerations for future research and agronomic applications. Front. Plant Sci. 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, D.L.; Edmond, C.; Harrington, J.L.; Nühse, T.S. Cell wall integrity controls root elongation via a general 1-aminocyclopropane-1-carboxylic acid-dependent, ethylene-independent pathway. Plant Physiol. 2011, 156, 596–604. [Google Scholar] [CrossRef] [Green Version]
- Dieryckx, C.; Gaudin, V.; Dupuy, J.W.; Bonneu, M.; Girard, V.; Job, D. Beyond plant defense: Insights on the potential of salicylic and methylsalicylic acid to contain growth of the phytopathogen Botrytis cinerea. Front. Plant Sci. 2015, 6, 859. [Google Scholar] [CrossRef] [Green Version]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Tugizimana, F.; Piater, L.A.; Steenkamp, P.A.; Madala, N.E.; Dubery, I.A. Untargeted metabolomics analysis reveals dynamic changes in azelaic acid- and salicylic acid derivatives in LPS-treated Nicotiana tabacum cells. Biochem. Biophys. Res. Commun. 2017, 482, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- Klessig, D.F.; Tian, M.; Choi, H.W. Multiple targets of salicylic acid and its derivatives in plants and animals. Front. Immunol. 2016, 7, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dean, J.V.; Mohammed, L.A.; Fitzpatrick, T. The formation, vacuolar localization, and tonoplast transport of salicylic acid glucose conjugates in tobacco cell suspension cultures. Planta 2005, 221, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Mareya, C.R.; Tugizimana, F.; Piater, L.A.; Madala, N.E.; Steenkamp, P.A.; Dubery, I.A. Untargeted metabolomics reveal defensome-related metabolic reprogramming in Sorghum bicolor against infection by Burkholderia andropogonis. Metabolites 2019, 9, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, P.G.; Dos Santos, F.N.; Zanotta, S.; Eberlin, M.N.; Carazzone, C. Metabolomics of Solanum lycopersicum infected with Phytophthora infestans leads to early detection of late blight in asymptomatic plants. Molecules 2018, 23, 3330. [Google Scholar] [CrossRef] [Green Version]
- Zeiss, D.R.; Piater, L.A.; Dubery, I.A. Hydroxycinnamate amides: Intriguing conjugates of plant protective metabolites. Trends Plant Sci. 2020, 26. [Google Scholar] [CrossRef]
- Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Metabolic profiling of PGPR-treated tomato plants reveal priming-related adaptations of secondary metabolites and aromatic amino acids. Metabolites 2020, 10, 210. [Google Scholar] [CrossRef]
- Shiman, S.A. Your Trusted Partner in Agriculturtal Remedies. Available online: http://ac.shimansa.co.za (accessed on 21 August 2020).
- Madala, N.E.; Tugizimana, F.; Steenkamp, P.A. Development and optimization of an UPLC-QTOF-MS/MS method based on an in-source collision induced dissociation approach for comprehensive discrimination of chlorogenic acids isomers from Momordica plant species. J. Anal. Methods Chem. 2014, 2014, 50879. [Google Scholar] [CrossRef]
- Ncube, E.N.; Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Dubery, I.A.; Madala, N.E. Analyses of chlorogenic acids and related cinnamic acid derivatives from Nicotiana tabacum tissues with the aid of UPLC-QTOF-MS/MS based on the in-source collision-induced dissociation method. Chem. Cent. J. 2014, 8, 66. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef] [Green Version]
- Dictionary of Natural Products. Available online: www.dnp.chemnetbase.com (accessed on 21 August 2020).
- Chemspider—Search and Share Chemistry. Available online: www.chemspider.com (accessed on 21 August 2020).
- MetaboAnalyst—Statistical, Functional and Integrative Analysis of Metabolomics Data. Available online: www.metaboanalyst.ca (accessed on 21 August 2020).
- Xia, J.; Mandal, R.; Sinelnikov, I.V.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—A comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, J.; Wishart, D.S. Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhlongo, M.I.; Piater, L.A.; Steenkamp, P.A.; Labuschagne, N.; Dubery, I.A. Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum lycopersicum Plants Responding to Phytophthora capsici. Metabolites 2020, 10, 466. https://doi.org/10.3390/metabo10110466
Mhlongo MI, Piater LA, Steenkamp PA, Labuschagne N, Dubery IA. Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum lycopersicum Plants Responding to Phytophthora capsici. Metabolites. 2020; 10(11):466. https://doi.org/10.3390/metabo10110466
Chicago/Turabian StyleMhlongo, Msizi I., Lizelle A. Piater, Paul A. Steenkamp, Nico Labuschagne, and Ian A. Dubery. 2020. "Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum lycopersicum Plants Responding to Phytophthora capsici" Metabolites 10, no. 11: 466. https://doi.org/10.3390/metabo10110466
APA StyleMhlongo, M. I., Piater, L. A., Steenkamp, P. A., Labuschagne, N., & Dubery, I. A. (2020). Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum lycopersicum Plants Responding to Phytophthora capsici. Metabolites, 10(11), 466. https://doi.org/10.3390/metabo10110466