Mouse Age Matters: How Age Affects the Murine Plasma Metabolome
Abstract
:1. Introduction
2. Results
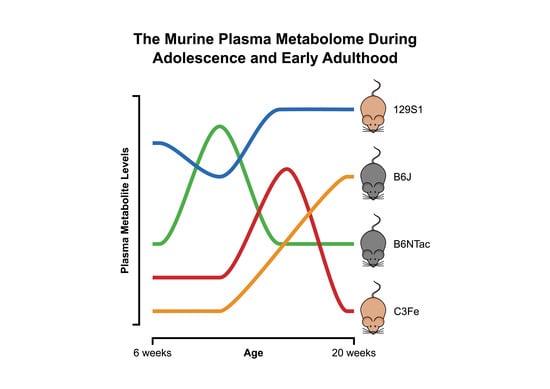
2.1. Metabolomic Changes during Adolescence and Early Adulthood
2.2. Metabolomic Similarities during Adolescence and Early Adulthood
2.3. Metabolomic Differences in Age-Matched Mouse Strains
3. Discussion
3.1. Metabolomic Changes during Adolescence and Early Adulthood
3.2. Strain-Specific Metabolomes during Adolescence and Early Adulthood
4. Materials and Methods
4.1. Mice
4.2. Plasma Sample Collection and Preparation
4.3. Targeted Metabolomics
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Bujak, R.; Struck-Lewicka, W.; Markuszewski, M.J.; Kaliszan, R. Metabolomics for laboratory diagnostics. J. Pharm. Biomed. Anal. 2015, 113, 108–120. [Google Scholar] [CrossRef] [PubMed]
- 2019 Report on the Statistics on the Use of Animals for Scientific Purposes in the Member States of the European Union in 2015–2017; European Commission: Brussels, Belgium, 2020; pp. 1–4.
- Dyar, K.A.; Lutter, D.; Artati, A.; Ceglia, N.J.; Liu, Y.; Armenta, D.; Jastroch, M.; Schneider, S.; de Mateo, S.; Cervantes, M.; et al. Atlas of Circadian Metabolism Reveals System-wide Coordination and Communication between Clocks. Cell 2018, 174, 1571–1585.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassett, S.A.; Young, W.; Fraser, K.; Dalziel, J.E.; Webster, J.; Ryan, L.; Fitzgerald, P.; Stanton, C.; Dinan, T.G.; Cryan, J.F.; et al. Metabolome and microbiome profiling of a stress-sensitive rat model of gut-brain axis dysfunction. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.L.; Kiersgaard, M.K.; Sørensen, D.B.; Mikkelsen, L.F. Fasting of mice: A review. Lab. Anim. 2013, 47, 225–240. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Thonusin, C.; Qi, N.R.; Burant, C.F.; Evans, C.R. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: Studies in a C57BL/6J mouse model. PLoS ONE 2015, 10, 1–19. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Cantó, C.; Jeninga, E.H.; Andreux, Ṕ.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The metabolic footprint of aging in mice. Sci. Rep. 2011, 1. [Google Scholar] [CrossRef]
- Tomás-Loba, A.; Bernardes de Jesus, B.; Mato, J.M.; Blasco, M.A. A metabolic signature predicts biological age in mice. Aging Cell 2013, 12, 93–101. [Google Scholar] [CrossRef]
- Kim, S.; Cheon, H.S.; Song, J.C.; Yun, S.M.; Park, S.I.; Jeon, J.P. Aging-related Changes in Mouse Serum Glycerophospholipid Profiles. Osong Public Health Res. Perspect. 2014, 5, 345–350. [Google Scholar] [CrossRef] [Green Version]
- Dutta, S.; Sengupta, P. Men and mice: Relating their ages. Life Sci. 2016, 152, 244–248. [Google Scholar] [CrossRef]
- Somerville, J.M.; Aspden, R.M.; Armour, K.E.; Armour, K.J.; Reid, D.M. Growth of C57Bl/6 Mice and the Material and Mechanical Properties of Cortical Bone from the Tibia. Calcif. Tissue Int. 2004, 74, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Pinchuk, L.M.; Filipov, N.M. Differential effects of age on circulating and splenic leukocyte populations in C57BL/6 and BALB/c male mice. Immun. Ageing 2008, 5, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, S.J.; Andrews, N.; Ball, D.; Bellantuono, I.; Gray, J.; Hachoumi, L.; Holmes, A.; Latcham, J.; Petrie, A.; Potter, P.; et al. Does age matter? The impact of rodent age on study outcomes. Lab. Anim. 2017, 51, 160–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brust, V.; Schindler, P.M.; Lewejohann, L. Lifetime development of behavioural phenotype in the house mouse (Mus musculus). Front. Zool. 2015, 12, S17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.; Dowling, A.R.; Marino, J.S.; Faulkner, L.D.; Bryant, B.; Brüning, J.C.; Elias, C.F.; Hill, J.W. Delayed Puberty but Normal Fertility in Mice With Selective Deletion of Insulin Receptors From Kiss1 Cells. Endocrinology 2013, 154, 1337–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chehab, F.F.; Mounzih, K.; Lu, R.; Lim, M.E. Early Onset of Reproductive Function in Normal Female Mice Treated with Leptin. Science 1997, 275, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Keri, R.A.; Lozada, K.L.; Abdul-Karim, F.W.; Nadeau, J.H.; Nilson, J.H. Luteinizing hormone induction of ovarian tumors: Oligogenic differences between mouse strains dictates tumor disposition. Proc. Natl. Acad. Sci. USA 2000, 97, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.F.; Karelus, K.; Felicio, L.S.; Johnson, T.E. Genetic Influences on the Timing of Puberty in Mice. Biol. Reprod. 1990, 42, 649–655. [Google Scholar] [CrossRef]
- Krewson, T.D.; Supelak, P.J.; Hill, A.E.; Singer, J.B.; Lander, E.S.; Nadeau, J.H.; Palmert, M.R. Chromosomes 6 and 13 Harbor Genes that Regulate Pubertal Timing in Mouse Chromosome Substitution Strains. Endocrinology 2004, 145, 4447–4451. [Google Scholar] [CrossRef] [Green Version]
- Moore, E.M.; Linsenbardt, D.N.; Melón, L.C.; Boehm, S.L., 2nd. Ontogenetic differences in adolescent and adult C57BL/6J and DBA/2J mice: Anxiety-like, locomotor, and consummatory behaviors. Dev. Psychobiol. 2011, 53, 141–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sisk, C.L.; Foster, D.L. The neural basis of puberty and adolescence. Nat. Neurosci. 2004, 7, 1040–1047. [Google Scholar] [CrossRef] [PubMed]
- Romeo, R.D.; Richardson, H.N.; Sisk, C.L. Puberty and the maturation of the male brain and sexual behavior: Recasting a behavioral potential. Neurosci. Biobehav. Rev. 2002, 26, 381–391. [Google Scholar] [CrossRef]
- Laviola, G.; Macrì, S.; Morley-Fletcher, S.; Adriani, W. Risk-taking behavior in adolescent mice: Psychobiological determinants and early epigenetic influence. Neurosci. Biobehav. Rev. 2003, 27, 19–31. [Google Scholar] [CrossRef]
- Bell, M.R. Comparing Postnatal Development of Gonadal Hormones and Associated Social Behaviors in Rats, Mice, and Humans. Endocrinology 2018, 159, 2596–2613. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, J.; Djukovic, D.; Daniel-MacDougall, C.; Gu, H.; Wu, X.; Chow, W.H. Metabolomics-identified metabolites associated with body mass index and prospective weight gain among Mexican American women. Obes. Sci. Pract. 2016, 2, 309–317. [Google Scholar] [CrossRef]
- Chuang, T.-L.; Lin, J.-W.; Wang, Y.-F. Bone Mineral Density as a Predictor of Atherogenic Indexes of Cardiovascular Disease, Especially in Nonobese Adults. Dis. Markers 2019, 2019, 1045098. [Google Scholar] [CrossRef] [Green Version]
- de Goede, K.E.; Harber, K.J.; Van den Bossche, J. Let’s Enter the Wonderful World of Immunometabolites. Trends Endocrinol. Metab. 2019, 30, 329–331. [Google Scholar] [CrossRef]
- Lilue, J.; Doran, A.G.; Fiddes, I.T.; Abrudan, M.; Armstrong, J.; Bennett, R.; Chow, W.; Collins, J.; Collins, S.; Czechanski, A.; et al. Sixteen diverse laboratory mouse reference genomes define strain-specific haplotypes and novel functional loci. Nat. Genet. 2018, 50, 1574–1583. [Google Scholar] [CrossRef]
- Timmermans, S.; Van Montagu, M.; Libert, C. Complete overview of protein-inactivating sequence variations in 36 sequenced mouse inbred strains. Proc. Natl. Acad. Sci. USA 2017, 114, 9158–9163. [Google Scholar] [CrossRef] [Green Version]
- Michaud, S.A.; Sinclair, N.J.; Pětrošová, H.; Palmer, A.L.; Pistawka, A.J.; Zhang, S.; Hardie, D.B.; Mohammed, Y.; Eshghi, A.; Richard, V.R.; et al. Molecular phenotyping of laboratory mouse strains using 500 multiple reaction monitoring mass spectrometry plasma assays. Commun. Biol. 2018, 1, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Leandro, J.; Violante, S.; Argmann, C.A.; Hagen, J.; Dodatko, T.; Bender, A.; Zhang, W.; Williams, E.G.; Bachmann, A.M.; Auwerx, J.; et al. Mild inborn errors of metabolism in commonly used inbred mouse strains. Mol. Genet. Metab. 2019, 126, 388–396. [Google Scholar] [CrossRef] [PubMed]
- Fujisaka, S.; Avila-Pacheco, J.; Soto, M.; Kostic, A.; Dreyfuss, J.M.; Pan, H.; Ussar, S.; Altindis, E.; Li, N.; Bry, L.; et al. Diet, Genetics, and the Gut Microbiome Drive Dynamic Changes in Plasma Metabolites. Cell Rep. 2018, 22, 3072–3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, Q.; Li, T.; Sun, J.; Liu, X.; Ren, J.; Fei, J. Metabolomic analysis of normal (C57BL/6J, 129S1/SvImJ) mice by gas chromatography–mass spectrometry: Detection of strain and gender differences. Talanta 2011, 85, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Mekada, K.; Abe, K.; Murakami, A.; Nakamura, S.; Nakata, H.; Moriwaki, K.; Obata, Y.; Yoshiki, A. Genetic differences among C57BL/6 substrains. Exp. Anim. 2009, 58, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Zurita, E.; Chagoyen, M.; Cantero, M.; Alonso, R.; González-Neira, A.; López-Jiménez, A.; López-Moreno, J.A.; Landel, C.P.; Benítez, J.; Pazos, F.; et al. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2011, 20, 481–489. [Google Scholar] [CrossRef]
- Römisch-Margl, W.; Prehn, C.; Bogumil, R.; Röhring, C.; Suhre, K.; Adamski, J. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 2012, 8, 133–142. [Google Scholar] [CrossRef]
- R Core Team. R A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]




| Mean Age | 129S1 | B6J | B6NTac | C3Fe | |||||
|---|---|---|---|---|---|---|---|---|---|
| (Weeks) | Female | Male | Female | Male | Female | Male | Female | Male | |
| n | 9 | 10 | 8 | 10 | 10 | 10 | 10 | 9 | |
| body weight (g) | 6 | 21.2 ± 1.4 | 22.5 ± 1.4 | 17.3 ± 1.0 | 21.8 ± 0.9 | 17.6 ± 1.2 | 19.8 ± 1.3 | 21.2 ± 1.5 | 24.1 ± 1.4 |
| age (d) | 42 ± 0 | 42 ± 0 | 46 ± 1 | 46 ± 0 | 42 ± 0 | 42 ± 0 | 44 ± 2 | 45 ± 3 | |
| n | 10 | 9 | 9 | 10 | 10 | 8 | 10 | 10 | |
| body weight (g) | 10 | 25.2 ± 3.5 | 27.5 ± 2.0 | 19.5 ± 0.8 | 26.4 ± 1.2 | 20.2 ± 1.3 | 25.1 ± 2.0 | 24.3 ± 1.6 | 28.0 ± 1.4 |
| age (d) | 70 ± 0 | 70 ± 0 | 74 ± 0 | 74 ± 0 | 70 ± 0 | 70 ± 0 | 72 ± 2 | 73 ± 3 | |
| weight gain (g/d) | 0.14 | 0.18 | 0.08 | 0.16 | 0.10 | 0.19 | 0.11 | 0.14 | |
| n | 10 | 10 | 9 | 9 | 10 | 9 | 9 | 9 | |
| body weight (g) | 14 | 26.9 ± 5.2 | 30.2 ± 2.2 | 21.0 ± 1.0 | 28.6 ± 1.1 | 21.8 ± 2.0 | 28.4 ± 2.3 | 27.2 ± 2.0 | 31.1 ± 1.5 |
| age (d) | 98 ± 0 | 98 ± 0 | 106 ± 4 | 104 ± 0 | 99 ± 0 | 98 ± 0 | 100 ± 2 | 101 ± 3 | |
| weight gain (g/d) | 0.06 | 0.09 | 0.05 | 0.07 | 0.06 | 0.12 | 0.10 | 0.11 | |
| n | 9 | 10 | 10 | 8 | 8 | 9 | 10 | 10 | |
| body weight (g) | 20 | 28.9 ± 6.3 | 32.5 ± 3.1 | 21.7 ± 0.9 | 29.9 ± 1.9 | 25.2 ± 3.4 | 32.4 ± 2.8 | 33.0 ± 2.6 | 36.1 ± 2.2 |
| age (d) | 133 ± 0 | 133 ± 0 | 147 ± 4 | 145 ± 0 | 140 ± 0 | 140 ± 0 | 151 ± 2 | 152 ± 3 | |
| weight gain (g/d) | 0.06 | 0.07 | 0.02 | 0.03 | 0.08 | 0.09 | 0.12 | 0.10 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pann, P.; de Angelis, M.H.; Prehn, C.; Adamski, J. Mouse Age Matters: How Age Affects the Murine Plasma Metabolome. Metabolites 2020, 10, 472. https://doi.org/10.3390/metabo10110472
Pann P, de Angelis MH, Prehn C, Adamski J. Mouse Age Matters: How Age Affects the Murine Plasma Metabolome. Metabolites. 2020; 10(11):472. https://doi.org/10.3390/metabo10110472
Chicago/Turabian StylePann, Patrick, Martin Hrabě de Angelis, Cornelia Prehn, and Jerzy Adamski. 2020. "Mouse Age Matters: How Age Affects the Murine Plasma Metabolome" Metabolites 10, no. 11: 472. https://doi.org/10.3390/metabo10110472
APA StylePann, P., de Angelis, M. H., Prehn, C., & Adamski, J. (2020). Mouse Age Matters: How Age Affects the Murine Plasma Metabolome. Metabolites, 10(11), 472. https://doi.org/10.3390/metabo10110472