Self-Redirection of Metabolic Flux toward Squalene and Ethanol Pathways by Engineered Yeast
Abstract
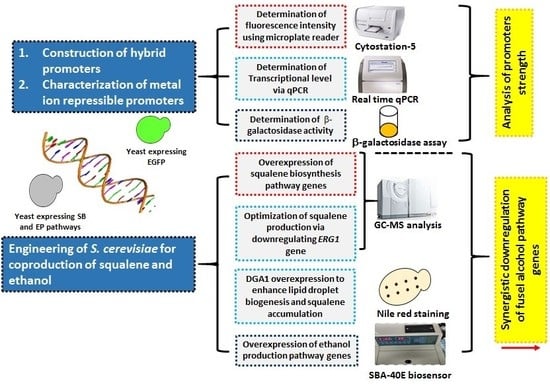
:1. Introduction
2. Results and Discussion
2.1. Tuning the Strength of Yeast Promoters
2.2. Characterization of Metal Ion Repressible Promoters
2.3. Overexpression of the Squalene Synthesis Pathway in Yeast Via Engineered Promoters
2.4. Optimization of Squalene Overproduction Via Overexpression of DGA1 and Downregulation of ERG1
2.5. Co-Overproduction of Ethanol in S. cerevisiae
2.6. Yeast Co-Producing the Squalene and Ethanol Self-Downregulated the Expression of Fusel Alcohol Pathway
3. Materials and Methods
3.1. Strains, Media, and Cells Cultivation
3.2. Tuning the Strength of Promoters
3.3. Characterization of Metal Ion Repressible Promoters
3.4. Characterization of the Strength of Engineered Promoters and Metal Ion Repressible Promoters
3.5. Yeast Engineering with Squalene and Ethanol Biosynthesis Pathways
3.6. Overexpression of the DGA1 in Engineered Strain
3.7. Downregulation of ERG1 Using Metal Ion Repressible Promoters
3.8. Total RNA Extraction and qRT-PCR Analysis
3.9. Nile Red Staining of Lipid Droplets in Engineered Strain
3.10. Extraction, Quantification and GC-MS Analysis of Squalene and Fusel Alcohols
3.11. Quantification of Ethanol
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, T.J. Squalene: Potential chemopreventive agent, Expert Opin. Investig. Drugs. 2000, 9, 1841–1848. [Google Scholar] [CrossRef]
- Nakazawa, A.; Kokubun, Y.; Matsuura, H.; Yonezawa, N.; Kose, R.; Yoshida, M.; Tanabe, Y.; Kusuda, E.; Van Thang, D.; Ueda, M.; et al. TLC screening of thraustochytrid strains for squalene production. J. Appl. Phycol. 2014, 26, 29–41. [Google Scholar] [CrossRef]
- Ahmed, M.S.; Ikram, S.; Rasool, A.; Li, C. Design and construction of short synthetic terminators for Β-amyrin production in Saccharomyces cerevisiae. Biochem. Eng. J. 2019, 105–116. [Google Scholar] [CrossRef]
- Kirby, J.; Romanini, D.W.; Paradise, E.M.; Keasling, J.D. Engineering triterpene production in Saccharomyces cerevisiae-β-amyrin synthase from Artemisia annua. FEBS J. 2008, 275, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Kohno, Y.; Egawa, Y.; Itoh, S.; Nagaoka, S.; Takahashi, M.; Mukai, K. Kinetic study of quenching reaction of singlet oxygen and scavenging reaction of free radical by squalene in n-butanol, Biochim. Biophys. Acta (BBA)/Lipids Lipid Metab. 1995, 1256, 52–56. [Google Scholar] [CrossRef]
- Amarowicz, R. Squalene: A natural antioxidant? Eur. J. Lipid Sci. Technol. 2009, 111, 411–412. [Google Scholar] [CrossRef]
- Aguilera, Y.; Dorado, M.E.; Prada, F.A.; Martínez, J.J.; Quesada, A.; Ruiz-Gutiérrez, V. The protective role of squalene in alcohol damage in the chick embryo retina. Exp. Eye Res. 2005, 80, 535–543. [Google Scholar] [CrossRef]
- Storm, H.M.; Oh, S.Y.; Kimler, B.F.; Norton, S. Radioprotection of mice by dietary squalene. Lipids 1993, 28, 555–559. [Google Scholar] [CrossRef]
- Hernández-Pérez, M.; Rabanal Gallego, R.M.; Pascual Alayón, P.J.; Brito Hernández, A. Squalene content in livers from deep-sea sharks caught in Canary Island waters. Mar. Freshw. Res. 1997, 48, 573–576. [Google Scholar] [CrossRef] [Green Version]
- Techera, E.J.; Klein, N. Sharks: Conservation, governance and management; Earthscan: London, UK, 2014. [Google Scholar] [CrossRef]
- Clarke, S. Shark product trade in Hong Kong and mainland China and implementation of the CITES shark listings; TRAFFIC East Asia: Hong Kong, China, 2004. [Google Scholar]
- Spanova, M.; Czabany, T.; Zellnig, G.N.; Leitner, E.; Hapala, I.; Daum, G.N. Effect of lipid particle biogenesis on the subcellular distribution of squalene in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 6127–6133. [Google Scholar] [CrossRef] [Green Version]
- Spanova, M.; Zweytick, D.; Lohner, K.; Klug, L.; Leitner, E.; Hermetter, A.; Daum, G. Influence of squalene on lipid particle/droplet and membrane organization in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta - Mol. Cell Biol. Lipids. 2012, 1821, 647–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasool, A.; Zhang, G.; Li, Z.; Li, C. Engineering of the terpenoid pathway in Saccharomyces cerevisiae co-overproduces squalene and the non-terpenoid compound oleic acid. Chem. Eng. Sci. 2016, 152, 457–467. [Google Scholar] [CrossRef]
- Rasool, A.; Ahmed, M.S.; Li, C. Overproduction of squalene synergistically downregulates ethanol production in Saccharomyces cerevisiae. Chem. Eng. Sci. 2016, 152, 370–380. [Google Scholar] [CrossRef]
- Hull, C.M.; Loveridge, E.J.; Rolley, N.J.; Donnison, I.S.; Kelly, S.L.; Kelly, D.E. Co-production of ethanol and squalene using a Saccharomyces cerevisiae ERG1 (squalene epoxidase) mutant and agro-industrial feedstock. Biotechnol. Biofuels. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nuñez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazelwood, L.A.; Daran, J.M.; Van Maris, A.J.A.; Pronk, J.T.; Dickinson, J.R. The Ehrlich pathway for fusel alcohol production: A century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008, 74, 2259–2266. [Google Scholar] [CrossRef] [Green Version]
- Ishida, N.; Saitoh, S.; Onishi, T.; Tokuhiro, K.; Nagamori, E.; Kitamoto, K.; Takahashi, H. The effect of pyruvate decarboxylase gene knockout in Saccharomyces cerevisiae on L-lactic acid production. Biosci. Biotechnol. Biochem. 2006. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Zhou, Y.J.; Huang, M.; Liu, Q.; Pereira, R.; David, F.; Nielsen, J. Reprogramming Yeast Metabolism from Alcoholic Fermentation to Lipogenesis. Cell 2018. [Google Scholar] [CrossRef] [Green Version]
- Koopman, F.; Beekwilder, J.; Crimi, B.; van Houwelingen, A.; Hall, R.D.; Bosch, D.; van Maris, A.J.A.; Pronk, J.T.; Daran, J.M. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2012. [Google Scholar] [CrossRef] [Green Version]
- Blazeck, J.; Garg, R.; Reed, B.; Alper, H.S. Controlling promoter strength and regulation in Saccharomyces cerevisiae using synthetic hybrid promoters. Biotechnol. Bioeng. 2012, 109, 2884–2895. [Google Scholar] [CrossRef]
- McNerney, M.P.; Styczynski, M.P. Precise control of lycopene production to enable a fast-responding, minimal-equipment biosensor. Metab. Eng. 2017. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Liang, F.; Lindberg, P. Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.M.; Garcia, D.E.; Redding-Johanson, A.M.; Friedland, G.D.; Chan, R.; Batth, T.S.; Haliburton, J.R.; Chivian, D.; Keasling, J.; Petzold, C.J.; et al. Optimization of a heterologous mevalonate pathway through the use of variant HMG-CoA reductases. Metab. Eng. 2011. [Google Scholar] [CrossRef]
- Segall-Shapiro, T.H.; Sontag, E.D.; Voigt, C.A. Engineered promoters enable constant gene expression at any copy number in bacteria. Nat. Biotechnol. 2018, 36, 352–358. [Google Scholar] [CrossRef]
- Johnson, A.O.; Gonzalez-Villanueva, M.; Tee, K.L.; Wong, T.S. An Engineered Constitutive Promoter Set with Broad Activity Range for Cupriavidus necator H16. ACS Synth. Biol. 2018, 7, 1918–1928. [Google Scholar] [CrossRef]
- Lee, K.S.; Kim, J.S.; Heo, P.; Yang, T.J.; Sung, Y.J.; Cheon, Y.; NKoo, H.M.; Yu, B.J.; Seo, J.H.; Jin, Y.S.; et al. Characterization of Saccharomyces cerevisiae promoters for heterologous gene expression in Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 2013. [Google Scholar] [CrossRef]
- Redden, H.; Alper, H.S. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015. [Google Scholar] [CrossRef] [Green Version]
- Biggs, B.W.; Lim, C.G.; Sagliani, K.; Shankar, S.; Stephanopoulos, G.; De Mey, M.; Ajikumar, P.K. Overcoming heterologous protein interdependency to optimize P450-mediated Taxol precursor synthesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2016. [Google Scholar] [CrossRef] [Green Version]
- Minkenberg, B.; Wheatley, M.; Yang, Y. CRISPR/Cas9-Enabled Multiplex Genome Editing and Its Application. Prog. Mol. Biol. Transl. Sci. 2017, 111–132. [Google Scholar] [CrossRef]
- Paradise, E.M.; Kirby, J.; Chan, R.; Keasling, J.D. Redirection of flux through the FPP branch-point in saccharomyces cerevisiae by down-regulating squalene synthase. Biotechnol. Bioeng. 2008, 100, 371–378. [Google Scholar] [CrossRef]
- Berthelet, S.; Usher, J.; Shulist, K.; Hamza, A.; Maltez, N.; Johnston, A.; Fong, Y.; Harris, L.J.; Baetz, K. Functional genomics analysis of the Saccharomyces cerevisiae iron responsive transcription factor Aft1 reveals iron-independent functions. Genetics 2010, 185, 1111–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dancis, A.; Haile, D.; Yuan, D.S.; Klausner, R.D. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 1994, 269, 25660–25667. [Google Scholar] [PubMed]
- Dai, Z.; Liu, Y.; Huang, L.; Zhang, X. Production of miltiradiene by metabolically engineered Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 2845–2853. [Google Scholar] [CrossRef] [PubMed]
- Westfall, P.J.; Pitera, D.J.; Lenihan, J.R.; Eng, D.; Woolard, F.X.; Regentin, R.; Horning, T.; Tsuruta, H.; Melis, D.J.; Owens, A.; et al. Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. USA 2012, 109. [Google Scholar] [CrossRef] [Green Version]
- Scalcinati, G.; Knuf, C.; Partow, S.; Chen, Y.; Maury, J.; Schalk, M.; Daviet, L.; Nielsen, J.; Siewers, V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene α-santalene in a fed-batch mode. Metab. Eng. 2012, 14, 91–103. [Google Scholar] [CrossRef]
- Moses, T.; Pollier, J.; Almagro, L.; Buyst, D.; Van Montagu, M.; Pedreño, M.A.; Martins, J.C.; Thevelein, J.M.; Goossens, A. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. USA 2014, 111, 1634–1639. [Google Scholar] [CrossRef] [Green Version]
- Albertsen, L.; Chen, Y.; Bach, L.S.; Rattleff, S.; Maury, J.; Brix, S.; Nielsen, J.; Mortensen, U.H. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2011, 77, 1033–1040. [Google Scholar] [CrossRef] [Green Version]
- BLOCH, K. Biological synthesis of cholesterol. Harvey Lect. 1952, 48, 68–88. [Google Scholar] [CrossRef]
- Donald, K.A.G.; Hampton, R.Y.; Fritz, I.B. Effects of overproduction of the catalytic domain of 3-hydroxy-3- methylglutaryl coenzyme A reductase on squalene synthesis in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1997, 63, 3341–3344. [Google Scholar] [CrossRef] [Green Version]
- Farhi, M.; Marhevka, E.; Masci, T.; Marcos, E.; Eyal, Y.; Ovadis, M.; Abeliovich, H.; Vainstein, A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab. Eng. 2011, 13, 474–481. [Google Scholar] [CrossRef]
- Venugopal, V.; Kumaran, A.K.; Sekhar Chatterjee, N.; Kumar, S.; Kavilakath, S.; Nair, J.R.; Mathew, S. Biochemical Characterization of Liver Oil of Echinorhinus brucus (Bramble Shark) and Its Cytotoxic Evaluation on Neuroblastoma Cell Lines (SHSY-5Y). Scientifica (Cairo) 2016. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.J.; Kwak, S.; Liu, J.J.; Lane, S.; Hua, Q.; Kweon, D.H.; Jin, Y.S. Improved squalene production through increasing lipid contents in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.L. Lipid droplet biogenesis. Curr. Opin. Cell Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Adeyo, O.; Horn, P.J.; Lee, S.K.; Binns, D.D.; Chandrahas, A.; Chapman, K.D.; Goodman, J.M. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radulovic, M.; Knittelfelder, O.; Cristobal-Sarramian, A.; Kolb, D.; Wolinski, H.; Kohlwein, S.D. The emergence of lipid droplets in yeast: Current status and experimental approaches. Curr. Genet. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, A.; Manneschmidt, A.; Dortch, R.; Verbruggie, R.; Dalhaimer, P. The Number of Lipid Droplets in the Fission Yeast S. pombe is a Function of the Cell Cycle Stage. Biophys. J. 2011. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.; Shi, B.; Ye, Z.; Li, X.; Liu, M.; Chen, Y.; Xia, J.; Nielsen, J.; Deng, Z.; Liu, T. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab. Eng. 2019. [Google Scholar] [CrossRef]
- Natunen, K.; Seppälä, J.; Schwenk, D.; Rischer, H.; Spilling, K.; Tamminen, T. Nile Red staining of phytoplankton neutral lipids: species-specific fluorescence kinetics in various solvents. J. Appl. Phycol. 2015. [Google Scholar] [CrossRef] [Green Version]
- Ruckenstuhl, C.; Lang, S.; Poschenel, A.; Eidenberger, A.; Baral, P.K.; Kohút, P.; Hapala, I.; Gruber, K.; Turnowsky, F. Characterization of squalene epoxidase of Saccharomyces cerevisiae by applying terbinafine-sensitive variants. Antimicrob. Agents Chemother. 2007. [Google Scholar] [CrossRef] [Green Version]
- Van Maris, A.J.A.; Geertman, J.M.A.; Vermeulen, A.; Groothuizen, M.K.; Winkler, A.A.; Piper, M.D.W.; Van Dijken, J.P.; Pronk, J.T. Directed Evolution of Pyruvate Decarboxylase-Negative Saccharomyces cerevisiae, Yielding a C2-Independent, Glucose-Tolerant, and Pyruvate-Hyperproducing Yeast. Appl. Environ. Microbiol. 2004. [Google Scholar] [CrossRef] [Green Version]
- Drewke, C.; Ciriacy, M. Overexpression, purification and properties of alcohol dehydrogenase IV from Saccharomyces cerevisiae. BBA - Gene Struct. Expr. 1988, 950, 54–60. [Google Scholar] [CrossRef]
- Raj, S.B.; Ramaswamy, S.; Plapp, B.V. Yeast Alcohol Dehydrogenase Structure and Catalysis. Biochemistry 2014, 53, 5791–5803. [Google Scholar] [CrossRef] [PubMed]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriksson, P.R.; Mendiratta, G.; McLaughlin, N.B.; Wolfsberg, T.G.; Marino-Ramirez, L.; Pompa, T.A.; Jainerin, M.; Landsman, D.; Shen, C.-H.; Clark, D.J. Global Regulation by the Yeast Spt10 Protein Is Mediated through Chromatin Structure and the Histone Upstream Activating Sequence Elements. Mol. Cell. Biol. 2005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arndt, K.; Fink, G.R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5′ TGACTC 3′ sequences. Proc. Natl. Acad. Sci. USA 1986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randise-Hinchliff, C.; Brickner, J.H. Transcription factors dynamically control the spatial organization of the yeast genome. Nucleus 2016. [Google Scholar] [CrossRef] [Green Version]
- Shao, Z.; Zhao, H.; Zhao, H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009. [Google Scholar] [CrossRef] [Green Version]
- Krogh, B.O.; Symington, L.S. Recombination Proteins in Yeast. Annu. Rev. Genet. 2004, 38, 233–271. [Google Scholar] [CrossRef] [Green Version]
- Jasin, M.; Rothstein, R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013. [Google Scholar] [CrossRef]
- Alemán-Nava, G.S.; Cuellar-Bermudez, S.P.; Cuaresma, M.; Bosma, R.; Muylaert, K.; Ritmann, B.E.; Parra, R. How to us Nile Red, a selective fluorescent stain for microalgal neutral lipids. J. Microbiol. Methods. 2016. [Google Scholar] [CrossRef]









© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manzoor, R.; Ahmed, M.; Riaz, N.; Kiani, B.H.; Kaleem, U.; Rashid, Y.; Nawaz, A.; Awan, M.U.F.; Khan, H.; Imtiaz, U.; et al. Self-Redirection of Metabolic Flux toward Squalene and Ethanol Pathways by Engineered Yeast. Metabolites 2020, 10, 56. https://doi.org/10.3390/metabo10020056
Manzoor R, Ahmed M, Riaz N, Kiani BH, Kaleem U, Rashid Y, Nawaz A, Awan MUF, Khan H, Imtiaz U, et al. Self-Redirection of Metabolic Flux toward Squalene and Ethanol Pathways by Engineered Yeast. Metabolites. 2020; 10(2):56. https://doi.org/10.3390/metabo10020056
Chicago/Turabian StyleManzoor, Robina, Maqbool Ahmed, Naveeda Riaz, Bushra Hafeez Kiani, Ullah Kaleem, Yasmeen Rashid, Ali Nawaz, Muhammad Umer Farooq Awan, Hooria Khan, Umera Imtiaz, and et al. 2020. "Self-Redirection of Metabolic Flux toward Squalene and Ethanol Pathways by Engineered Yeast" Metabolites 10, no. 2: 56. https://doi.org/10.3390/metabo10020056
APA StyleManzoor, R., Ahmed, M., Riaz, N., Kiani, B. H., Kaleem, U., Rashid, Y., Nawaz, A., Awan, M. U. F., Khan, H., Imtiaz, U., Rasheed, Y., Kaleem, I., & Rasool, A. (2020). Self-Redirection of Metabolic Flux toward Squalene and Ethanol Pathways by Engineered Yeast. Metabolites, 10(2), 56. https://doi.org/10.3390/metabo10020056