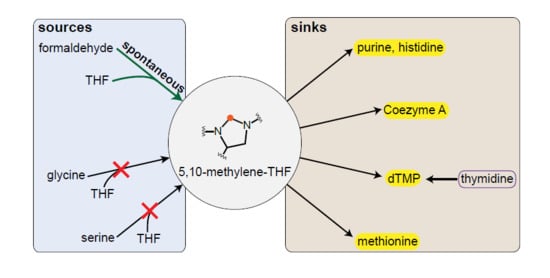
In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Strains and Genomic Modifications
4.2. Media and Growth Conditions
4.3. Formaldehyde Concentration Measurement
4.4. Isotopic-labeling Experiments
4.5. Flux Balance Analysis
- Reactions POR5 and GLYCK were removed from the model since they are not present in E. coli cells.
- The stoichiometry of the reaction THD2pp was changed, as the translocation uses only one proton instead of two.
- PFL and OBTFL were removed, since they are catalyzed by oxygen sensitive enzymes and therefore not active in aerobic conditions.
- Based on gathered evidence, FTHFLi was removed. This reaction is especially important in our case, since it could allow charging of THF with formate directly.
- We changed the directionality of three reactions in the nucleotide biosynthesis pathway (FTHFD, GARFT, and AICART) to be irreversible, in order to avoid nucleotides degradation to charge THF, since that is very unlikely to occur.
- We changed the thymidine periplasmic symporter reaction (THYMt3pp) to be reversible, based on our experimental evidence (adding thymidine to the media rescued growth in certain conditions).
- We knocked out the glycine cleavage system reaction and the GlyA reaction (GLYCL and GHMT2r).
- We added the formaldehyde-THF spontaneous condensation (THFSPONT) reaction: thf_c + fald_c -> mlthf_c + h2o_c.
- Removed the following reactions to avoid potential carbon fixation cycles: GLXCL, GLYCL, THRD, THRA2, THRA, SERD_L.
- Added NAD-dependent formate dehydrogenase (FDH), enabling the utilization of the formate that is created from formaldehyde as source of reducing power.
- Added the crotonoyl-CoA carboxylase reaction (EtMaCoA), required for the ethylmalonyl-CoA cycle: b2coa_c + nadph_c + co2_c + q8_c + h2o_c -> glx_c + ppcoa_c + nadp_c + q8h2_c.
- Added the propionyl-CoA carboxylase reaction (MMCDr), as required for recycling the propionyl-CoA, which was created by EtMaCoA: ppcoa_c + atp_c + hco3_c -> mmcoa__S_c + adp_c + h_c + pi_c.
- Made the methylmalonyl-CoA mutase reaction (MMM) reversible so that the methylmalonyl-CoA (mmcoa__S_c) can be recycled.
- Added the reactions of the serine cycle:
- ○
- serine-hydroxypyruvate amino-transferase (SGAT1): ser__L_c + akg_c -> glu__L_c + hpyr_c.
- ○
- glycine-glyoxylate amino-transferase (SGAT2): gly_c + akg_c -> glx_c + glu__L_c.
- ○
- malate thiokinase (MTK): mal__L_c + atp_c + coa_c -> malylcoa_c + adp_c + pi_c.
- ○
- malyl-CoA lyase (MCL): malylcoa_c -> accoa_c + glx_c.
4.6. Folate Profiling
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Khersonsky, O.; Tawfik, D.S. Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010, 79, 471–505. [Google Scholar] [CrossRef]
- Bar-Even, A.; Salah Tawfik, D. Engineering specialized metabolic pathways-is there a room for enzyme improvements? Curr. Opin. Biotechnol. 2013, 24, 310–319. [Google Scholar] [CrossRef]
- Delauney, A.J.; Verma, D.P.S. Proline Biosynthesis and Osmoregulation in Plants. Plant J. 1993, 4, 215–223. [Google Scholar] [CrossRef]
- Dekant, W.; Vamvakas, S. Glutathione-dependent bioactivation of xenobiotics. Xenobiotica 1993, 23, 873–887. [Google Scholar] [CrossRef]
- Metz, B.; Kersten, G.F.; Hoogerhout, P.; Brugghe, H.F.; Timmermans, H.A.; de Jong, A.; Meiring, H.; ten Hove, J.; Hennink, W.E.; Crommelin, D.J.; et al. Identification of formaldehyde-induced modifications in proteins: Reactions with model peptides. J. Biol. Chem. 2004, 279, 6235–6243. [Google Scholar] [CrossRef] [Green Version]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic methylotrophic prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Springer: Berlin, Heidelberg, Germany, 2013; pp. 267–285. [Google Scholar]
- Crowther, G.J.; Kosaly, G.; Lidstrom, M.E. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 2008, 190, 5057–5062. [Google Scholar] [CrossRef] [Green Version]
- Vorholt, J.A.; Marx, C.J.; Lidstrom, M.E.; Thauer, R.K. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J. Bacteriol. 2000, 182, 6645–6650. [Google Scholar] [CrossRef] [Green Version]
- Blakley, R.L. The reaction of tetrahydropteroylglutamic acid and related hydropteridines with formaldehyde. Biochem. J. 1959, 72, 707–715. [Google Scholar] [CrossRef] [Green Version]
- Large, P.J.; Quayle, J.R. Microbial growth on C1 compounds. 5. Enzyme activities in extracts of Pseudomonas AM1. Biochem. J. 1963, 87, 386–396. [Google Scholar] [CrossRef] [Green Version]
- Heptinstall, J.; Quayle, J.R. Pathways leading to and from serine during growth of Pseudomonas AM1 on C1 compounds or succinate. Biochem. J. 1970, 117, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Marx, C.J.; Van Dien, S.J.; Lidstrom, M.E. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 2005, 3, e16. [Google Scholar] [CrossRef] [Green Version]
- Kallen, R.G.; Jencks, W.P. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J. Biol. Chem. 1966, 241, 5851–5863. [Google Scholar]
- Dev, I.K.; Harvey, R.J. A complex of N5,N10-methylenetetrahydrofolate dehydrogenase and N5,N10-methenyltetrahydrofolate cyclohydrolase in Escherichia coli. Purification, subunit structure, and allosteric inhibition by N10-formyltetrahydrofolate. J. Biol. Chem. 1978, 253, 4245–4253. [Google Scholar]
- Powers, S.G.; Snell, E.E. Ketopantoate hydroxymethyltransferase. II. Physical, catalytic, and regulatory properties. J. Biol. Chem. 1976, 251, 3786–3793. [Google Scholar]
- Dev, I.K.; Yates, B.B.; Leong, J.; Dallas, W.S. Functional role of cysteine-146 in Escherichia coli thymidylate synthase. Proc. Natl. Acad. Sci. USA 1988, 85, 1472–1476. [Google Scholar] [CrossRef] [Green Version]
- Guenther, B.D.; Sheppard, C.A.; Tran, P.; Rozen, R.; Matthews, R.G.; Ludwig, M.L. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat. Struct. Biol. 1999, 6, 359–365. [Google Scholar] [CrossRef]
- Neidhardt, F.C.; Ingraham, J.L.; Schaechter, M. Building blocks needed to produce 1 g of E. coli protoplasm. In Physiology of the Bacterial Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 1990; pp. 134–143. [Google Scholar]
- Maden, B.E. Tetrahydrofolate and tetrahydromethanopterin compared: Functionally distinct carriers in C1 metabolism. Biochem. J. 2000, 350, 609–629. [Google Scholar] [CrossRef]
- Yishai, O.; Goldbach, L.; Tenenboim, H.; Lindner, S.N.; Bar-Even, A. Engineered Assimilation of Exogenous and Endogenous Formate in Escherichia coli. ACS Synth. Biol. 2017, 6, 1722–1731. [Google Scholar] [CrossRef] [Green Version]
- He, H.; Edlich-Muth, C.; Lindner, S.N.; Bar-Even, A. Ribulose Monophosphate Shunt Provides Nearly All Biomass and Energy Required for Growth of E. coli. ACS Synth. Biol. 2018, 7, 1601–1611. [Google Scholar] [CrossRef]
- Hassan-Abdallah, A.; Zhao, G.; Chen, Z.W.; Mathews, F.S.; Schuman Jorns, M. Arginine 49 is a bifunctional residue important in catalysis and biosynthesis of monomeric sarcosine oxidase: A context-sensitive model for the electrostatic impact of arginine to lysine mutations. Biochemistry 2008, 47, 2913–2922. [Google Scholar] [CrossRef]
- Jensen, S.I.; Lennen, R.M.; Herrgard, M.J.; Nielsen, A.T. Seven gene deletions in seven days: Fast generation of Escherichia coli strains tolerant to acetate and osmotic stress. Sci. Rep. 2015, 5, 17874. [Google Scholar] [CrossRef] [Green Version]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006–2008. [Google Scholar] [CrossRef] [Green Version]
- Guzman, E.C.; Martin, C.M. Thymineless death, at the origin. Front. Microbiol. 2015, 6, 499. [Google Scholar] [CrossRef] [Green Version]
- Sugantino, M.; Zheng, R.; Yu, M.; Blanchard, J.S. Mycobacterium tuberculosis ketopantoate hydroxymethyltransferase: Tetrahydrofolate-independent hydroxymethyltransferase and enolization reactions with alpha-keto acids. Biochemistry 2003, 42, 191–199. [Google Scholar] [CrossRef]
- Contestabile, R.; Paiardini, A.; Pascarella, S.; di Salvo, M.L.; D’Aguanno, S.; Bossa, F. L-Threonine aldolase, serine hydroxymethyltransferase and fungal alanine racemase. A subgroup of strictly related enzymes specialized for different functions. Eur. J. Biochem. 2001, 268, 6508–6525. [Google Scholar] [CrossRef]
- Monk, J.M.; Lloyd, C.J.; Brunk, E.; Mih, N.; Sastry, A.; King, Z.; Takeuchi, R.; Nomura, W.; Zhang, Z.; Mori, H.; et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat. Biotechnol. 2017, 35, 904–908. [Google Scholar] [CrossRef]
- Edwards, J.S.; Ramakrishna, R.; Palsson, B.O. Characterizing the metabolic phenotype: A phenotype phase plane analysis. Biotechnol. Bioeng. 2002, 77, 27–36. [Google Scholar] [CrossRef]
- Milo, R.; Jorgensen, P.; Moran, U.; Weber, G.; Springer, M. BioNumbers—The database of key numbers in molecular and cell biology. Nucleic Acids Res. 2010, 38, D750–D753. [Google Scholar] [CrossRef] [Green Version]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Fact. 2013, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Flegr, J. Two Distinct Types of Natural Selection in Turbidostat-like and Chemostat-like Ecosystems. J. Theor. Biol. 1997, 188, 121–126. [Google Scholar] [CrossRef]
- Thomason, L.C.; Costantino, N.; Court, D.L.E. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. 2007, 79, 1–17. [Google Scholar] [CrossRef]
- Imlay, J.A. The mismetallation of enzymes during oxidative stress. J. Biol. Chem. 2014, 289, 28121–28128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nash, T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction. Biochem. J. 1953, 55, 416–421. [Google Scholar] [CrossRef] [Green Version]
- You, L.; Page, L.; Feng, X.; Berla, B.; Pakrasi, H.B.; Tang, Y.J. Metabolic pathway confirmation and discovery through 13C-labeling of proteinogenic amino acids. J. Vis. Exp. 2012, e3583. [Google Scholar] [CrossRef] [Green Version]
- Giavalisco, P.; Li, Y.; Matthes, A.; Eckhardt, A.; Hubberten, H.M.; Hesse, H.; Segu, S.; Hummel, J.; Kohl, K.; Willmitzer, L. Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J. 2011, 68, 364–376. [Google Scholar] [CrossRef]
- Ebrahim, A.; Lerman, J.A.; Palsson, B.O.; Hyduke, D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst. Biol. 2013, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Parra, P.A.; Urrea-López, R.; Díaz de la Garza, R.I. Folate analysis in complex food matrices: Use of a recombinant Arabidopsis γ-glutamyl hydrolase for folate deglutamylation. Food Res. Int. 2013, 54, 177–185. [Google Scholar] [CrossRef]
- Ramos-Parra, P.A.; García-Salinas, C.; Hernández-Brenes, C.; Díaz de la Garza, R.I. Folate Levels and Polyglutamylation Profiles of Papaya (Carica papaya cv. Maradol) during Fruit Development and Ripening. J. Agric. Food Chem. 2013, 61, 3949–3956. [Google Scholar] [CrossRef]






| Strain | Genotype | Source |
|---|---|---|
| SIJ488 | K-12 MG1655 Tn7::para-exo-beta-gam; prha-FLP; xylSpm-IsceI | [23] |
| DH5α | F- endA1 glnV44 thi-1 recA1 relA1 gyrA96 deoR nupG purB20 φ80dlacZΔM15 Δ(lacZYA-argF)U169, hsdR17(rK-mK+), λ- | Lab collection |
| Frm | SIJ488 ΔfrmRAB | [21] |
| gcv_don | K-12 MG1655 ΔgcvTHP::Km | Lab collection |
| ltaE_don | K-12 BW25113 ΔltaE::Km | [24] |
| panB_don | K-12 BW25113 ΔpanB::Km | [24] |
| glyA_don | K-12 BW25113 ΔglyA::Km | [24] |
| C1AΔ3 | SIJ488 ΔfrmRAB ΔgcvTHP ΔglyA::Km | This study |
| C1AΔ5 | SIJ488 ΔfrmRAB ΔgcvTHP ΔltaE ΔpanB ΔglyA::Km | This study |
| Plasmid | Genes | Source |
| pZASS | p15A ori; StrepR; Ppgi-20 | Lab collection |
| pZASS-soxA | pZASS::soxA; sarcosine oxidase | [21] |
| pZASS-CgadhA | pZASS::CgadhA; methanol dehydrogenase | [21] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, H.; Noor, E.; Ramos-Parra, P.A.; García-Valencia, L.E.; Patterson, J.A.; Díaz de la Garza, R.I.; Hanson, A.D.; Bar-Even, A. In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate. Metabolites 2020, 10, 65. https://doi.org/10.3390/metabo10020065
He H, Noor E, Ramos-Parra PA, García-Valencia LE, Patterson JA, Díaz de la Garza RI, Hanson AD, Bar-Even A. In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate. Metabolites. 2020; 10(2):65. https://doi.org/10.3390/metabo10020065
Chicago/Turabian StyleHe, Hai, Elad Noor, Perla A. Ramos-Parra, Liliana E. García-Valencia, Jenelle A. Patterson, Rocío I. Díaz de la Garza, Andrew D. Hanson, and Arren Bar-Even. 2020. "In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate" Metabolites 10, no. 2: 65. https://doi.org/10.3390/metabo10020065
APA StyleHe, H., Noor, E., Ramos-Parra, P. A., García-Valencia, L. E., Patterson, J. A., Díaz de la Garza, R. I., Hanson, A. D., & Bar-Even, A. (2020). In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate. Metabolites, 10(2), 65. https://doi.org/10.3390/metabo10020065