Wood Metabolomic Responses of Wild and Cultivated Grapevine to Infection with Neofusicoccum parvum, a Trunk Disease Pathogen
Abstract
:1. Introduction
2. Results
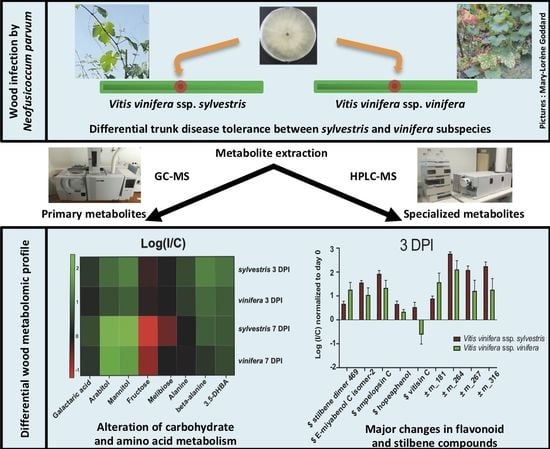
2.1. Changes in Primary Metabolites in Wood upon N. parvum Inoculation
2.2. Primary Metabolic Pathways Impacted by N. parvum Infection
2.3. N. parvum Inoculation Resulted in More Intense Primary Metabolism Alterations in V. v. subsp. sylvestris Compared to V. v. subsp. vinifera
2.4. Changes in Specialized Metabolites in Wood upon N. parvum Inoculation
2.5. Response to N. parvum in V. v. subsp. sylvestris Is Characterized by Higher Induction of Resveratrol Oligomer Levels after Wood Inoculation
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Fungal Material and Wood Inoculation
4.3. Metabolite Extraction
4.4. GC-MS Analysis
4.5. HPLC-DAD-MS Analysis
4.6. Metabolomic Data Processing and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hofstetter, V.; Buyck, B.; Croll, D.; Viret, O.; Couloux, A.; Gindro, K. What if esca disease of grapevine were not a fungal disease? Fungal Divers. 2012, 54, 51–67. [Google Scholar] [CrossRef] [Green Version]
- Bertsch, C.; Ramírez-Suero, M.; Magnin-Robert, M.; Larignon, P.; Chong, J.; Mansour, E.A.; Spagnolo, A.; Clement, C.; Fontaine, F. Grapevine trunk diseases: Complex and still poorly understood. Plant Pathol. 2012, 62, 243–265. [Google Scholar] [CrossRef] [Green Version]
- Ahimera, N.; Driever, G.F.; Michailides, T.J. Relationships among propagule numbers of Botryosphaeria dothidea, latent infections, and severity of panicle and shoot blight in Pistachio orchards. Plant Dis. 2003, 87, 846–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbes-Torres, J.R. The status of Botryosphaeriaceae species infecting grapevines. Phytopathol. Mediterr. 2011, 50, 5–45. [Google Scholar] [CrossRef]
- Larignon, P. Maladies Cryptogamiques Du Bois De La Vigne: Symptomatologie Et Agents Pathogènes. Available online: http://www.vignevin.com/menu-haut/actualites/article.html?tx_ttnews%5Btt_news%5D=368&tx_ttnews%5BbackPid%5D=918&cHash=2c0eccd030 (accessed on 19 January 2017).
- Fontaine, F.; Pinto, C.; Vallet, J.; Clément, C.; Gomes, A.C.; Spagnolo, A. The effects of grapevine trunk diseases (GTDs) on vine physiology. Eur. J. Plant Pathol. 2015, 144, 707–721. [Google Scholar] [CrossRef]
- Úrbez-Torres, J.R.; Gubler, W.D. Pathogenicity of Botryosphaeriaceae species isolated from grapevine cankers in California. Plant Dis. 2009, 93, 584–592. [Google Scholar] [CrossRef] [Green Version]
- Mehl, J.W.M.; Slippers, B.; Roux, J.; Wingfield, M.J. Cankers and other diseases caused by the Botryosphaeriaceae. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CABI Publishing: Warringford, UK, 2013; pp. 298–317. [Google Scholar]
- Slippers, B.; Wingfield, M.J. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: Diversity, ecology and impact. Fungal Biol. Rev. 2007, 21, 90–106. [Google Scholar] [CrossRef]
- Larignon, P.; Fulchic, R.; Cere, L.; Dubos, B. Observation on Black Dead Arm in French vineyards. Phytopathol. Mediterr. 2001, 40, 336–342. [Google Scholar]
- Evidente, A.; Punzo, B.; Andolfi, A.; Cimmino, A.; Melck, D.; Luque, J. Lipophilic phytotoxins produced by Neofusicoccum parvum, a grapevine canker agent. Phytopathol. Mediterr. 2010, 49, 74–79. [Google Scholar]
- Mansour, E.A.; Debieux, J.-L.; Ramírez-Suero, M.; Bénard-Gellon, M.; Magnin-Robert, M.; Spagnolo, A.; Chong, J.; Farine, S.; Bertsch, C.; L’Haridon, F.; et al. Phytotoxic metabolites from Neofusicoccum parvum, a pathogen of Botryosphaeria dieback of grapevine. Phytochemistry 2015, 115, 207–215. [Google Scholar] [CrossRef]
- Burruano, S.; Giambra, S.; Mondello, V.; Dellagreca, M.; Basso, S.; Tuzi, A.; Andolfi, A. Naphthalenone polyketides produced by Neofusicoccum parvum, a fungus associated with grapevine Botryosphaeria dieback. Phytopathol. Mediterr. 2016, 55, 197–206. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Adrian, M.; Trouvelot, S.; Spagnolo, A.; Jacquens, L.; Letousey, P.; Rabenoelina, F.; Harir, M.; Roullier-Gall, C.; Clément, C.; et al. Alterations in grapevine leaf metabolism occur prior to esca apoplexy appearance. Mol. Plant Microbe Interact. 2017, 30, 946–959. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Felgueiras, M.; Graça, G.; Rodrigues, J.E.; Barros, A.; Gil, A.M.; Dias, A.C.P. NMR metabolomics of esca disease-affected Vitis vinifera cv. Alvarinho leaves. J. Exp. Bot. 2010, 61, 4033–4042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camps, C.; Kappel, C.; Lecomte, P.; Léon, C.; Gomès, E.; Coutos-Thévenot, P.; Delrot, S. A transcriptomic study of grapevine (Vitis vinifera cv. Cabernet-Sauvignon) interaction with the vascular ascomycete fungus Eutypa lata. J. Exp. Bot. 2010, 61, 1719–1737. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Felgueiras, M.; Cunha, A.; Chicau, G.; Ferreres, F.; Dias, A.C.P. Differential phenolic production in leaves of Vitis vinifera cv. Alvarinho affected with esca disease. Plant Physiol. Biochem. 2017, 112, 45–52. [Google Scholar] [CrossRef]
- Moret, F.; Lemaître-Guillier, C.; Grosjean, C.; Clément, G.; Coelho, C.; Negrel, J.; Jacquens, L.; Morvan, G.; Mouille, G.; Trouvelot, S.; et al. Clone-dependent expression of esca disease Revealed by leaf metabolite analysis. Front. Plant Sci. 2019, 9, 9. [Google Scholar] [CrossRef]
- Magnin-Robert, M.; Spagnolo, A.; Boulanger, A.; Joyeux, C.; Clément, C.; Mansour, E.A.; Fontaine, F. Changes in plant metabolism and accumulation of fungal metabolites in response to esca proper and apoplexy expression in the whole grapevine. Phytopathology 2016, 106, 541–553. [Google Scholar] [CrossRef]
- Rusjan, D.; Persic, M.; Likar, M.; Biniari, K.; Mikulic-Petkovsek, M. Phenolic responses to esca-associated fungi in differently decayed grapevine woods from different trunk parts of ‘Cabernet Sauvignon’. J. Agric. Food Chem. 2017, 31, 6615–6624. [Google Scholar] [CrossRef]
- Grosman, J.; Doublet, B. Maladies du bois de la vigne. Synthèse des dispositifs d’observation au vignoble, de l’observatoire 2003-2008 au réseau d’épidémiosurveillance actuel. Phytoma 2012, 651, 31–35. [Google Scholar]
- Bruez, E.; Lecomte, P.; Grosman, J.; Doublet, B.; Bertsch, C.; Fontaine, F.; Ugaglia, A.; Teissedre, P.-L.; Da Costa, J.-P.; Guerin-Dubrana, L.; et al. Overview of grapevine trunk diseases in France in the 2000s. Phytopathol. Mediterr. 2013, 52, 262–275. [Google Scholar]
- Guan, X.; Essakhi, S.; Laloue, H.; Nick, P.; Bertsch, C.; Chong, J. Mining new resources for grape resistance against Botryosphaeriaceae: A focus on Vitis vinifera subsp. Sylvestris. Plant Pathol. 2015, 65, 273–284. [Google Scholar] [CrossRef]
- Schymanski, E.L.; Jeon, J.; Gulde, R.; Fenner, K.; Ruff, M.; Singer, H.P.; Hollender, J. Identifying small molecules via high resolution mass spectrometry: Communicating confidence. Environ. Sci. Technol. 2014, 48, 2097–2098. [Google Scholar] [CrossRef]
- Lai, Z.; Tsugawa, H.; Wohlgemuth, G.; Mehta, S.; Mueller, M.; Zheng, Y.; Ogiwara, A.; Meissen, J.; Showalter, M.; Takeuchi, K.; et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods 2017, 15, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Tsugawa, H.; Kind, T.; Nakabayashi, R.; Yukihira, D.; Tanaka, W.; Cajka, T.; Saito, K.; Fiehn, O.; Arita, M. Hydrogen rearrangement rules: Computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem. 2016, 88, 7946–7958. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, N.D.; Watrous, J.; A Kapono, C.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Flamini, R.; De Rosso, M. Chapter 5-High-Resolution mass spectrometry and biological properties of grapevine and wine stilbenoids. In Studies in Natural Products Chemistry; Rahman, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 175–210. [Google Scholar]
- Gabaston, J.; Leborgne, C.; Waffo-Teguo, P.; Valls, J.; Pinto, A.P.; Richard, T.; Cluzet, S.; Merillon, J.-M. Wood and roots of major grapevine cultivars and rootstocks: A comparative analysis of stilbenes by UHPLC-DAD-MS/MS and NMR. Phytochem. Anal. 2019, 30, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Moss, R.; Mao, Q.; Taylor, D.; Saucier, C. Investigation of monomeric and oligomeric wine stilbenoids in red wines by ultra-high-performance liquid chromatography/electrospray ionization quadrupole time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2013, 27, 1815–1827. [Google Scholar] [CrossRef]
- Oshima, Y.; Ueno, Y.; Hikino, H.; Ling-Ling, Y.; Kun-Ying, Y. Ampelopsins A, B and C, new oligostilbenes of VAR. Tetrahedron 1990, 46, 5121–5126. [Google Scholar] [CrossRef]
- Chaher, N.; Arraki, K.; Dillinseger, E.; Temsamani, H.; Bernillon, S.; Pedrot, E.; Delaunay, J.-C.; Mérillon, J.-M.; Monti, J.-P.; Izard, J.-C.; et al. Bioactive stilbenes from Vitis vinifera grapevine shoots extracts. J. Sci. Food Agric. 2013, 94, 951–954. [Google Scholar] [CrossRef]
- Billet, K.; Houillé, B.; De Bernonville, T.D.; Besseau, S.; Oudin, A.; Courdavault, V.; Delanoue, G.; Guérin, L.; Clastre, M.; Giglioli-Guivarc’H, N.; et al. Field-based metabolomics of Vitis vinifera L. stems provides new insights for genotype discrimination and polyphenol metabolism structuring. Front. Plant Sci. 2018, 9, 798. [Google Scholar] [CrossRef]
- Daldoul, S.; Boubakri, H.; Gargouri, M.; Mliki, A. Recent advances in biotechnological studies on wild grapevines as valuable resistance sources for smart viticulture. Mol. Biol. Rep. 2020, 47, 3141–3153. [Google Scholar] [CrossRef] [PubMed]
- Duan, D.; Halter, D.; Baltenweck, R.; Tisch, C.; Tröster, V.; Kortekamp, A.; Hugueney, P.; Nick, P. Genetic diversity of stilbene metabolism in Vitis sylvestris. J. Exp. Bot. 2015, 66, 3243–3257. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.-K.; Ahn, S.; Cho, H.Y.; Yun, H.Y.; Park, J.H.; Lim, J.; Lee, J.; Kwon, S.W. Metabolic response induced by parasitic plant-fungus interactions hinder amino sugar and nucleotide sugar metabolism in the host. Sci. Rep. 2016, 6, 37434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.-L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [Green Version]
- Cardot, C.; Mappa, G.; La Camera, S.; Gaillard, C.; Vriet, C.; Lecomte, P.; Ferrari, G.; Coutos-Thévenot, P. Comparison of the molecular responses of tolerant, susceptible and highly susceptible grapevine cultivars during interaction with the pathogenic fungus Eutypa lata. Front. Plant Sci. 2019, 10, 991. [Google Scholar] [CrossRef] [Green Version]
- Lima, M.; Machado, A.F.; Gubler, W.D. Metabolomic study of Chardonnay grapevines double-stressed with Esca-associated fungi and drought. Phytopathology 2017, 107, 669–680. [Google Scholar] [CrossRef]
- Zulak, K.G.; Weljie, A.M.; Vogel, H.J.; Facchini, P.J. Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures. BMC Plant Biol. 2008, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Broeckling, C.D.; Huhman, D.; Farag, M.A.; Smith, J.T.; May, G.D.; Mendes, P.; Dixon, R.A.; Sumner, L.W. Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 2004, 56, 323–336. [Google Scholar] [CrossRef] [Green Version]
- Monselise, E.B.-I.; Parola, A.H.; Kost, D. Low-frequency electromagnetic fields induce a stress effect upon higher plants, as evident by the universal stress signal, alanine. Biochem. Biophys. Res. Commun. 2003, 302, 427–434. [Google Scholar] [CrossRef]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef] [Green Version]
- Seifikalhor, M.; Aliniaeifard, S.; Hassani, B.; Niknam, V.; Lastochkina, O. Diverse role of γ-aminobutyric acid in dynamic plant cell responses. Plant Cell Rep. 2019, 38, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Mead, O.; Thynne, E.; Winterberg, B.; Solomon, P.S. Characterising the role of GABA and its metabolism in the wheat pathogen Stagonospora nodorum. PLoS ONE 2013, 8, e78368. [Google Scholar] [CrossRef] [PubMed]
- Jobic, C.; Boisson, A.-M.; Gout, E.; Rascle, C.; Fevre, M.; Cotton, P.; Bligny, R. Metabolic processes and carbon nutrient exchanges between host and pathogen sustain the disease development during sunflower infection by Sclerotinia sclerotiorum. Planta 2007, 226, 251–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dulermo, T.; Rascle, C.; Chinnici, G.; Gout, E.; Bligny, R.; Cotton, P. Dynamic carbon transfer during pathogenesis of sunflower by the necrotrophic fungus Botrytis cinerea: From plant hexoses to mannitol. New Phytol. 2009, 183, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Conde, A.; Regalado, A.; Rodrigues, D.; Costa, J.M.; Blumwald, E.; Chaves, M.M.; Geros, H. Polyols in grape berry: Transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 2014, 66, 889–906. [Google Scholar] [CrossRef] [Green Version]
- Conde, A.; Silva, P.; Agasse, A.; Conde, C.; Geros, H. Mannitol transport and mannitol dehydrogenase activities are coordinated in Olea europaea under salt and osmotic stresses. Plant Cell Physiol. 2011, 52, 1766–1775. [Google Scholar] [CrossRef] [Green Version]
- Meena, M.; Prasad, V.; Zehra, A.; Gupta, V.K.; Upadhyay, R.S. Mannitol metabolism during pathogenic fungal–host interactions under stressed conditions. Front. Microbiol. 2015, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Poutaraud, A.; Hugueney, P. Metabolism and roles of stilbenes in plants. Plant Sci. 2009, 177, 143–155. [Google Scholar] [CrossRef]
- Amalfitano, C.; Agrelli, D.; Arrigo, A.; Mugnai, L.; Surico, G.; Evidente, A. Stilbene polyphenols in the brown red wood of Vitis vinifera cv. Sangiovese affected by esca proper. Phytopathol. Mediterr. 2011, 50, 224–235. [Google Scholar] [CrossRef]
- Stempien, E.; Goddard, M.-L.; Leva, Y.; Bénard-Gellon, M.; Laloue, H.; Farine, S.; Kieffer-Mazet, F.; Tarnus, C.; Bertsch, C.; Chong, J. Secreted proteins produced by fungi associated with Botryosphaeria dieback trigger distinct defense responses in Vitis vinifera and Vitis rupestris cells. Protoplasma 2017, 255, 613–628. [Google Scholar] [CrossRef]
- Amalfitano, C.; Peduto, F.; Mugnai, L.; Evidente, A.; Surico, G. Chemical characterisation of stilbenic polyphenols from esca diseased wood and their role in defence mechanisms. Phytopathol. Mediterr. 2005, 44, 99. [Google Scholar]
- Lambert, C.; Khiook, I.L.K.; Lucas, S.; Télef-Micouleau, N.; Mérillon, J.-M.; Cluzet, S. A Faster and a stronger defense response: One of the key elements in grapevine explaining its lower level of susceptibility to esca? Phytopathology 2013, 103, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-W.; Sun, K.; Guan, Y.-X.; Qiu, M.-H.; Zhang, L.; Dai, C.-C. Fungal endophyte Phomopsis liquidambari biodegrades soil resveratrol: A potential allelochemical in peanut monocropping systems. J. Sci. Food Agric. 2019, 99, 5899–5909. [Google Scholar] [CrossRef] [PubMed]
- Lambert, C.; Bisson, J.; Waffo-Téguo, P.; Papastamoulis, Y.; Richard, T.; Corio-Costet, M.-F.; Mérillon, J.-M.; Cluzet, S. Phenolics and their antifungal role in grapevine wood decay: Focus on the Botryosphaeriaceae Family. J. Agric. Food Chem. 2012, 60, 11859–11868. [Google Scholar] [CrossRef]
- Stempien, E.; Goddard, M.-L.; Wilhelm, K.; Tarnus, C.; Bertsch, C.; Chong, J. Grapevine Botryosphaeria dieback fungi have specific aggressiveness factor repertory involved in wood decay and stilbene metabolization. PLoS ONE 2017, 12, e0188766. [Google Scholar] [CrossRef]
- Nick, P. Schützen und nützen-Von der erhaltung zur anwendung. fallbeispiel europäische wildrebe. Hoppea Denkschr. Regensb. Bot. Ges. 2014, 159–173. Available online: https://www.botanik.kit.edu/garten/downloads/Pub_Nick_2014_sylvestris.pdf (accessed on 3 June 2020).
- Noctor, G.; Bergot, G.; Mauve, C.; Thominet, D.; Lelarge-Trouverie, C.; Prioul, J.L. A comparative study of amino acid measurement in leaf extracts by gas chromatography-time of flight-mass spectrometry and high performance liquid chromatography with fluorescence detection. Metabolomics 2007, 3, 161–174. [Google Scholar] [CrossRef]
- Press, H.; Teukolsky, S.A. Savitzky-Golay smoothing filters. Comput. Phys. 1990, 4, 669. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and integrative metabolomics data analysis. Curr. Protoc. Bioinform. 2019, 68, 86. [Google Scholar] [CrossRef]
- Van Den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van Der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cumming, G.; Fidler, F.; Vaux, D.L. Error bars in experimental biology. J. Cell Biol. 2007, 177, 7–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Zhao, X.; Lü, X.; Lin, X.; Xu, G. A data preprocessing strategy for metabolomics to reduce the mask effect in data analysis. Front. Mol. Biosci. 2015, 2, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Labois, C.; Wilhelm, K.; Laloue, H.; Tarnus, C.; Bertsch, C.; Goddard, M.-L.; Chong, J. Wood Metabolomic Responses of Wild and Cultivated Grapevine to Infection with Neofusicoccum parvum, a Trunk Disease Pathogen. Metabolites 2020, 10, 232. https://doi.org/10.3390/metabo10060232
Labois C, Wilhelm K, Laloue H, Tarnus C, Bertsch C, Goddard M-L, Chong J. Wood Metabolomic Responses of Wild and Cultivated Grapevine to Infection with Neofusicoccum parvum, a Trunk Disease Pathogen. Metabolites. 2020; 10(6):232. https://doi.org/10.3390/metabo10060232
Chicago/Turabian StyleLabois, Clément, Kim Wilhelm, Hélène Laloue, Céline Tarnus, Christophe Bertsch, Mary-Lorène Goddard, and Julie Chong. 2020. "Wood Metabolomic Responses of Wild and Cultivated Grapevine to Infection with Neofusicoccum parvum, a Trunk Disease Pathogen" Metabolites 10, no. 6: 232. https://doi.org/10.3390/metabo10060232
APA StyleLabois, C., Wilhelm, K., Laloue, H., Tarnus, C., Bertsch, C., Goddard, M.-L., & Chong, J. (2020). Wood Metabolomic Responses of Wild and Cultivated Grapevine to Infection with Neofusicoccum parvum, a Trunk Disease Pathogen. Metabolites, 10(6), 232. https://doi.org/10.3390/metabo10060232