Potential Lipid Signatures for Diagnosis and Prognosis of Sepsis and Systemic Inflammatory Response Syndrome
Abstract
:1. Introduction
2. Results
2.1. Subject and Clinical Data
2.2. Analysis of Plasma Samples
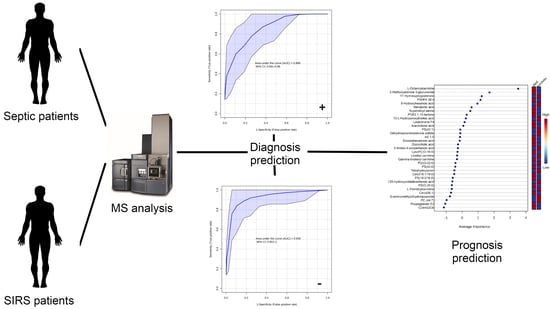
2.2.1. Analysis of Lipid Signatures for Diagnosis
2.2.2. Performance Evaluation of Diagnostic Lipid Signatures Used for Prognostic Prediction
2.2.3. Performance Evaluation of L-Octanoylcarnitine as Diagnostic and Prognostic Predictor
3. Discussion
4. Materials and Methods
4.1. Study Groups
4.2. Sample Collection, Preparation and LC-MS/MS Analysis
4.2.1. LC-MS Analysis
4.2.2. Data Acquisition and Preprocessing
4.3. Statistical Analysis
4.3.1. Exploratory Analysis
4.3.2. Analysis of Biomarkers for Diagnosis
4.3.3. Putative Identification of Lipids and Metabolomics Pathway Analysis
4.3.4. Performance Evaluation of Diagnostic Biomarkers Used for Prognostic Prediction
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SIRS | systemic inflammatory response syndrome |
| SOFA | sequential organ failure assessment |
| qSOFA | quick SOFA |
| APACHE evaluation | acute physiology and chronic health |
| PCT | procalcitonin |
| SD | standard deviation |
| BMI | body mass index |
| CRP | C-reactive protein |
| SAPS | simplified acute physiology score |
| COPD | chronic obstructive pulmonary disease |
| AP | arterial pressure |
| AKI | acute kidney injury |
| mmHg | millimeters of mercury |
| mg | milligram |
| dL | deciliter |
| INR | international normalized ratio |
| mm3 | cubic millimeter |
| PaO2 | partial pressure of oxygen |
| FiO2 | fraction of inspired oxygen |
| ICU | intensive care unit |
| UTI | urinary tract infection |
| USF | Universidade São Francisco |
| QC | quality control |
| PCA | principal component analysis |
| AUC | area under curve |
| ROC | receiver operating characteristic |
| RF | random forest |
| MSI | Metabolomics Standards Initiative |
| QC-RFSC correction | quality control random forest based signal |
| IQR | interquartile range |
| MCCV | Monte Carlo cross-validation |
| MS | mass spectrometry |
| HMDB | Human Metabolome Database |
| PC | phosphatidylcholine |
| PG | phosphatidylglycerol |
| ANOVA | analysis of variance |
| UPLC | ultra performance liquid chromatography |
| ACN | MS data-independent acquisition |
| MSE | n acetonitrile |
| EDTA | ethylenediamine tetraacetic acid |
| QTOF | quadrupole time-of-flight mass spectrometry |
| FAHFA | fatty acid esters of hydroxy fatty acids |
| TCA | tricarboxylic acid cycle |
| GSL | glycosphigolipids |
| LC-MS | liquid chromatography–mass spectrometry |
References
- Singer, M.; Deutschman, C.S.; Seymour, C.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA J. Am. Med. Assoc. 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Kyriacou, D.N. Government Regulation of Sepsis Care. JAMA 2019, 322, 250–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shankar-Hari, M.; Harrison, D.A.; Rowan, K.M. Differences in Impact of Definitional Elements on Mortality Precludes International Comparisons of Sepsis Epidemiology—A Cohort Study Illustrating the Need for Standardized Reporting. Crit. Care Med. 2016, 44, 2223–2230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivers, E.P.; Coba, V.; Visbal, A.; Whitmill, M.; Amponsah, D. Management of Sepsis: Early Resuscitation. Clin. Chest Med. 2008, 29, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.M.; Dobb, G.J.; Knuiman, M.; Finn, J.; Lee, K.Y.; Webb, S.A. A comparison of admission and worst 24-hour Acute Physiology and Chronic Health Evaluation II scores in predicting hospital mortality: A retrospective cohort study. Crit. Care 2005, 10, R4. [Google Scholar] [CrossRef] [Green Version]
- Metnitz, P.G.; Moreno, R.P.; Almeida, E.; Jordan, B.; Bauer, P.; Campos, R.A.; Iapichino, G.; Edbrooke, D.; Capuzzo, M.; Le Gall, J.R. SAPS 3-From evaluation of the patient to evaluation of the intensive care unit. Part 1: Objectives, methods and cohort description. Intensive Care Med. 2005, 31, 1336–1344. [Google Scholar] [CrossRef] [Green Version]
- Larsen, F.F.; Petersen, J.A. Novel biomarkers for sepsis: A narrative review. Eur. J. Intern. Med. 2017, 45, 46–50. [Google Scholar] [CrossRef]
- Pregernig, A.; Müller, M.; Held, U.; Beck-Schimmer, B. Prediction of mortality in adult patients with sepsis using six biomarkers: A systematic review and meta-analysis. Ann. Intensive Care 2019, 9, 125. [Google Scholar] [CrossRef]
- Pool, R.; Gomez, H.; Kellum, J.A. Mechanisms of Organ Dysfunction in Sepsis. Crit. Care Clin. 2018, 34, 63–80. [Google Scholar] [CrossRef]
- Churpek, M.M.; Zadravecz, F.J.; Winslow, C.; Howell, M.D.; Edelson, D.P. Incidence and Prognostic Value of the Systemic Inflammatory Response Syndrome and Organ Dysfunctions inWard Patients. Am. J. Respir. Crit. Care Med. 2015, 192, 958–964. [Google Scholar] [CrossRef] [Green Version]
- Gando, S.; Shiraishi, A.; Abe, T.; Kushimoto, S.; Mayumi, T.; Fujishima, S.; Hagiwara, A.; Shiino, Y.; Shiraishi, S.i.; Hifumi, T.; et al. The SIRS criteria have better performance for predicting infection than qSOFA scores in the emergency department. Sci. Rep. 2020, 10, 8095. [Google Scholar] [CrossRef] [PubMed]
- Olivier, M.; Asmis, R.; Hawkins, G.A.; Howard, T.D.; Cox, L.A. The Need for Multi-Omics Biomarker Signatures in Precision Medicine. Int. J. Mol. Sci. 2019, 20, 4781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, J.; Zhang, L.; Yan, F.; Wang, X. Clinical lipidomics: A new way to diagnose human diseases. Clin. Transl. Med. 2018, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Clémot, M.; Sênos Demarco, R.; Jones, D.L. Lipid Mediated Regulation of Adult Stem Cell Behavior. Front. Cell Dev. Biol. 2020, 8, 115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubler, M.J.; Kennedy, A.J. Role of lipids in the metabolism and activation of immune cells. J. Nutr. Biochem. 2016, 34, 1–7. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, V.B.; Ekroos, K.; Liebisch, G.; Wakelam, M. Lipidomics: Current state of the art in a fast moving field. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1466. [Google Scholar] [CrossRef]
- Mecatti, G.C.; Fernandes Messias, M.C.; Sant’Anna Paiola, R.M.; Figueiredo Angolini, C.F.; da Silva Cunha, I.B.; Eberlin, M.N.; de Oliveira Carvalho, P. Lipidomic Profiling of Plasma and Erythrocytes From Septic Patients Reveals Potential Biomarker Candidates. Biomark. Insights 2018, 13, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mecatti, G.C.; Messias, M.C.F.; de Oliveira Carvalho, P. Lipidomic profile and candidate biomarkers in septic patients. Lipids Health Dis. 2020, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Huang, Y.; Zhu, Y.; Xia, L.; Wang, R.; Xiao, K.; Wang, H.; Yan, P.; Wen, B.; Cao, L.; et al. Discrimination of sepsis stage metabolic profiles with an LC/MS-MS-based metabolomics approach. BMJ Open Respir. Res. 2016, 1, e000056. [Google Scholar] [CrossRef] [Green Version]
- Ferrario, M.; Cambiaghi, A.; Brunelli, L.; Giordano, S.; Caironi, P.; Guatteri, L.; Raimondi, F.; Gattinoni, L.; Latini, R.; Masson, S.; et al. Mortality prediction in patients with severe septic shock: A pilot study using a target metabolomics approach. Sci. Rep. 2016, 6, 20391. [Google Scholar] [CrossRef]
- Ludwig, K.R.; Hummon, A.B. Mass spectrometry for the discovery of biomarkers of sepsis. Mol. Biosyst. 2017, 13, 648–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, K.; Kum, C.K. How to Appraise a Prognostic Study. World J. Surg. 2005, 29, 567–569. [Google Scholar] [CrossRef]
- Sinapidis, D.; Kosmas, V.; Vittoros, V.; Koutelidakis, I.M.; Pantazi, A.; Stefos, A.; Katsaros, K.E.; Akinosoglou, K.; Bristianou, M.; Toutouzas, K.; et al. Progression into sepsis: An individualized process varying by the interaction of comorbidities with the underlying infection. BMC Infect. Dis. 2018, 18, 242. [Google Scholar] [CrossRef] [Green Version]
- Broadhurst, D.; Goodacre, R.; Reinke, S.N.; Kuligowski, J.; Wilson, I.D.; Lewis, M.R.; Dunn, W.B. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018, 14, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Triba, M.N.; Amathieu, R.; Lin, X.; Bouchemal, N.; Hantz, E.; Le Moyec, L.; Savarin, P. Nuclear magnetic resonance-based serum metabolomic analysis reveals different disease evolution profiles between septic shock survivors and non-survivors. Crit. Care 2019, 23, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Angelo, G.; Capasso, S.; Sticco, L.; Russo, D. Glycosphingolipids: Synthesis and functions. FEBS J. 2013, 280, 6338–6353. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, H.; Nagafuku, M.; Suzuki, A.; Iwabuchi, K.; Inokuchi, J. The regulatory roles of glycosphingolipid-enriched lipid rafts in immune systems. FEBS Lett. 2018, 592, 3921–3942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inokuchi, J.I.; Inamori, K.I.; Kabayama, K.; Nagafuku, M.; Uemura, S.; Go, S.; Suzuki, A.; Ohno, I.; Kanoh, H.; Shishido, F. Biology of GM3 Ganglioside. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar] [CrossRef]
- Sonnino, S.; Prinetti, A. Membrane Domains and the “Lipid Raft” Concept. Curr. Med. Chem. 2012, 20, 4–21. [Google Scholar] [CrossRef]
- Schmerler, D.; Neugebauer, S.; Ludewig, K.; Bremer-Streck, S.; Brunkhorst, F.M.; Kiehntopf, M. Targeted metabolomics for discrimination of systemic inflammatory disorders in critically ill patients. J. Lipid Res. 2012, 53, 1369–1375. [Google Scholar] [CrossRef] [Green Version]
- Neugebauer, S.; Giamarellos-Bourboulis, E.J.; Pelekanou, A.; Marioli, A.; Baziaka, F.; Tsangaris, I.; Bauer, M.; Kiehntopf, M. Metabolite Profiles in Sepsis. Crit. Care Med. 2016, 44, 1649–1662. [Google Scholar] [CrossRef]
- Fruhwirth, G.O.; Loidl, A.; Hermetter, A. Oxidized phospholipids: From molecular properties to disease. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2007, 1772, 718–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sassoli, C.; Pierucci, F.; Zecchi-Orlandini, S.; Meacci, E. Sphingosine 1-Phosphate (S1P)/S1P Receptor Signaling and Mechanotransduction: Implications for Intrinsic Tissue Repair/Regeneration. Int. J. Mol. Sci. 2019, 20, 5545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunz, T.C.; Kozjak-Pavlovic, V. Diverse Facets of Sphingolipid Involvement in Bacterial Infections. Front. Cell Dev. Biol. 2019, 7, 203. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, Y.; Teng, S.; Li, K. Prediction of sepsis mortality using metabolite biomarkers in the blood: A meta-analysis of death-related pathways and prospective validation. BMC Med. 2020, 18, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Feng, Y.W.; Yao, Y.M. Potential therapy strategy: Targeting mitochondrial dysfunction in sepsis. Mil. Med. Res. 2018, 5, 41. [Google Scholar] [CrossRef] [Green Version]
- Houten, S.M.; Wanders, R.J.A. A general introduction to the biochemistry of mitochondrial fatty acid b-oxidation. J. Inherit. Metab. Dis. 2010, 33, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Jung, S.; Lee, S.H.; Lee, J.H. Association between Arterial Stiffness and Serum L-Octanoylcarnitine and Lactosylceramide in Overweight Middle-Aged Subjects: 3-Year Follow-Up Study. PLoS ONE 2015, 10, e0119519. [Google Scholar] [CrossRef]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef]
- Zoni, E.; Minoli, M.; Bovet, C.; Wehrhan, A.; Piscuoglio, S.; Ng, C.K.Y.; Gray, P.C.; Spahn, M.; Thalmann, G.N.; Kruithof-de Julio, M. Preoperative plasma fatty acid metabolites inform risk of prostate cancer progression and may be used for personalized patient stratification. BMC Cancer 2019, 19, 1216. [Google Scholar] [CrossRef]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a Class of Endogenous Mammalian Lipids with Anti-Diabetic and Anti-inflammatory Effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef] [Green Version]
- B Gowda, S.G.; Fuda, H.; Tsukui, T.; Chiba, H.; Hui, S.P. Discovery of Eicosapentaenoic Acid Esters of Hydroxy Fatty Acids as Potent Nrf2 Activators. Antioxidants 2020, 9, 397. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Moraes-Vieira, P.M.; Castoldi, A.; Aryal, P.; Yee, E.U.; Vickers, C.; Parnas, O.; Donaldson, C.J.; Saghatelian, A.; Kahn, B.B. Branched Fatty Acid Esters of Hydroxy Fatty Acids (FAHFAs) Protect against Colitis by Regulating Gut Innate and Adaptive Immune Responses. J. Biol. Chem. 2016, 291, 22207–22217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Audano, M.; Maldini, M.; De Fabiani, E.; Mitro, N.; Caruso, D. Gender-related metabolomics and lipidomics: From experimental animal models to clinical evidence. J. Proteom. 2018, 178, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Bone, R.C.; Balk, R.A.; Cerra, F.B.; Dellinger, R.P.; Fein, A.M.; Knaus, W.A.; Schein, R.M.; Sibbald, W.J. Definitions for Sepsis and Organ Failure and Guidelines for the Use of Innovative Therapies in Sepsis. Chest 1992, 101, 1644–1655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, A.M.A.; Messias, M.C.; Duarte, G.H.; de Santis, G.K.; Mecatti, G.C.; Porcari, A.M.; Murgu, M.; Simionato, A.V.C.; Rocha, T.; Martinez, C.A.; et al. Plasma Lipid Profile Reveals Plasmalogens as Potential Biomarkers for Colon Cancer Screening. Metabolites 2020, 10, 262. [Google Scholar] [CrossRef]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.H.; Russell, D.W.; et al. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [Green Version]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef]
- Luan, H.; Ji, F.; Chen, Y.; Cai, Z. statTarget: A streamlined tool for signal drift correction and interpretations of quantitative mass spectrometry-based omics data. Anal. Chim. Acta 2018, 1036, 66–72. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Luan, H.; Ji, F.; Chen, Y.; Cai, Z. Quality control-based signal drift correction and interpretations of metabolomics/proteomics data using random forest regression. Biorxiv 2018. [Google Scholar] [CrossRef] [Green Version]
- Smilde, A.K.; van der Werf, M.J.; Bijlsma, S.; van der Werff-van der Vat, B.J.C.; Jellema, R.H. Fusion of Mass Spectrometry-Based Metabolomics Data. Anal. Chem. 2005, 77, 6729–6736. [Google Scholar] [CrossRef] [PubMed]
- Hackstadt, A.J.; Hess, A.M. Filtering for increased power for microarray data analysis. BMC Bioinform. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [Green Version]






| Sepsis | SIRS | Sepsis vs. SIRS | |||||
|---|---|---|---|---|---|---|---|
| N | Mean | SD | N | Mean | SD | p-Value | |
| Age | 21 | 55.52 | 19.79 | 21 | 48.00 | 17.44 | 0.20 |
| BMI | 21 | 25.10 | 4.92 | 21 | 24.96 | 3.49 | 0.92 |
| SAPSIII | 21 | 56.95 | 17.17 | 21 | 53.05 | 14.72 | 0.43 |
| Risk of death (%) | 21 | 38.33 | 29.39 | 21 | 29.03 | 24.01 | 0.27 |
| SOFA score | 21 | 5.14 | 2.95 | 21 | 6.43 | 3.11 | 0.18 |
| Comorbidities | |||||||
| Systemic hypertension | 7 | 0.33 | - | 1 | 0.05 | - | 0.05 |
| Diabetes mellitus | 5 | 0.24 | - | 0 | 0.00 | - | 0.05 |
| Dyslipidemia | 0 | 0.00 | - | 0 | 0.00 | - | 1.00 |
| Coronary insufficiency | 1 | 0.48 | - | 0 | 0.00 | - | 1.00 |
| COPD | 4 | 0.19 | - | 0 | 0.00 | - | 0.11 |
| Neoplasm | 3 | 0.14 | - | 0 | 0.00 | - | 0.23 |
| Organ dysfunction | |||||||
| by patient | 21 | 2.05 | - | 21 | 2.05 | - | 0.82 |
| AP < 90 mmHg | 10 | 0.48 | - | 16 | 0.76 | - | 0.11 |
| Lactate > 20 mg/dL | 11 | 0.52 | - | 13 | 0.62 | - | 0.76 |
| AKI | 5 | 0.24 | - | 6 | 0.29 | - | 1.00 |
| Total bilirubin > 2 mg/dL | 3 | 0.14 | - | 1 | 0.05 | - | 0.61 |
| INR > 1.6 | 8 | 0.38 | - | 1 | 0.05 | - | 0.02 |
| Platelets < 150,000/mm3 | 1 | 0.05 | - | 4 | 19.05 | - | 0.34 |
| PaO2/FiO2 ratio < 300 | 5 | 0.24 | - | 1 | 4.76 | - | 0.18 |
| Site of infection | |||||||
| Pneumonia | 7 | 0.33 | - | - | - | - | - |
| Abdominal | 9 | 0.43 | - | - | - | - | - |
| UTI | 1 | 0.05 | - | - | - | - | - |
| Others | 4 | 0.19 | - | - | - | - | - |
| Outcome (death) | |||||||
| ICU length of stay | 21 | 7.91 | 5.99 | 21 | 10.81 | 6.90 | 0.15 |
| Total outcome | 7 | 0.33 | - | 7 | 0.33 | - | 1.00 |
| Measured m/z | Ion Mode | Adducts | Lipid Assignment | Proposed Formula | Mass Error (ppm) | Abundance Sepsis | Abundance SIRS |
|---|---|---|---|---|---|---|---|
| 129.0555 | - | M-H2O-H [1−] | Mevalonic acid a | C6H12O4 | −1.69 | 1385.62 (722.52) | 1277.46 (743.76) |
| 132.0657 | + | M+H [1+] | 2-amino-4-oxopentanoic acid a | C5H9NO3 | 1.51 | 991.28 (466.89) | 1013.26 (580.62) |
| 133.0854 | + | M+H [1+] | 6-hydroxyhexanoic acid a | C6H12O3 | −3.76 | 679.72 (866.08) | 782.53 (914.21) |
| 238.1169 | + | M+H [1+] | S-aminomethyldihydrolipoamide a | C9H20N2OS2 | 0.39 | 1114.93 (320.52) | 1135.13 (536.29) |
| 282.1251 | - | M−2H [2−] | Leukotriene F4 b | C28H44N2O8S | −2.54 | 1154.50 (1659.83) | 1066.37 (662.07) |
| 288.2181 | + | M+H [1+] | L-octanoylcarnitine b | C15H29NO4 | 4.16 | 1452.21 (1113.79) | 659.59 (430.61) |
| 293.2119 | - | M−H2O-H [1−] | 13-L-hydroperoxylinoleic acid b | C18H32O4 | −1.08 | 891.92 (644.89) | 751.29 (539.68) |
| 295.2277 | - | M−H [1−] | 13S-hydroxyoctadecadienoic acid b | C18H32O3 | −0.68 | 909.30 (645.14) | 492.81 (355.81) |
| 303.2333 | - | M−H [1−] | Arachidonic acid b | C20H32O2 | 0.99 | 817.75 (713.45) | 1104.24 (653.40) |
| 326.2670 | + | M+H−H2O [+1] | N-palmitoyl serine a | C19H37NO4 | −5.64 | 2286.56 (4267.87) | 3535.59 (5678.59) |
| 327.2332 | - | M−H [1−] | Docosahexaenoic acid a | C22H32O2 | 0.61 | 812.39 (508.09) | 780.96 (365.68) |
| 331.2280 | + | M+H [1+] | 17-hydroxyprogesterone b | C21H30O3 | 3.62 | 1142.74 (400.95) | 1133.59 (530.18) |
| 335.2218 | + | M+H [1+] | PGE2 1,15-lactone a | C20H30O4 | 0.30 | 709.67 (263.66) | 561.39 (187.29) |
| 353.2326 | + | M+H [+1] | Prostaglandin E2 b | C20H32O5 | 1.13 | 1405.19 (1305.15) | 698.16 (637.36) |
| 367.1578 | - | M−H [1−] | Dehydroepiandrosterone sulfate b | C19H28O5S | −1.90 | 614.23 (463.79) | 2599.17 (1954.71) |
| 397.2051 | - | M−H2O-H [−1] | 7-[(2,4,6-trihydroxy-2,5,5,8a-tetramethyl-decahydronaphthalen-1-yl)methoxy]-2H-chromen-2-one a | C24H32O6 | 7.28 | 436.61 (267.23) | 1948.12 (1895.83) |
| 400.3438 | + | M+H [1+] | L-palmitoylcarnitine b | C23H45NO4 | 4.25 | 1275.23 (1117.99) | 684.14 (547.16) |
| 422.3260 | + | M+H [1+] | Gamma-linolenyl carnitine a | C25H43NO4 | −1.18 | 994.63 (388.35) | 832.62 (254.73) |
| 424.3432 | + | M+H [1+] | Linoleyl carnitine b | C25H45NO4 | 2.59 | 1087.23 (1266.79) | 598.35 (520.68) |
| 426.3589 | + | M+ACN+H [+1] | Tetrahydropersin a | C23H44O4 | 2.89 | 1146.89 (1099.49) | 568.64 (508.05) |
| 464.3016 | - | M-H [1−] | Glycocholic acid a | C26H43NO6 | −0.43 | 1506.15 (4407.32) | 639.15 (1149.97) |
| 477.2132 | + | M+H [1+] | 2-methoxyestrone 3-glucuronide a | C25H32O9 | 2.72 | 1057.75 (267.34) | 1088.04 (224.43) |
| 510.3940 | + | M+H [1+] | LysoPC (O-18:0) b | C8H20NO6PR | 4.31 | 932.19 (572.29) | 776.95 (514.12) |
| 557.4584 | − | M−H [−1] | FAHFA 36:4 a | C36H62O4 | 1.62 | 760.47 (665.67) | 1528.53 (1306.92) |
| 582.5110 | − | M+FA−H [−1] | Cer (d16:1/18:0) a | C34H67NO3 | 1.38 | 1931.34 (2267.96) | 593.15 (508.32) |
| 610.5423 | − | M+FA−H [−1] | Cer (d36:1) a | C36H71NO3 | 1.32 | 1497.42 (786.64) | 707.04 (471.59) |
| 753.5293 | − | M+FA−H [−1] | PG (O−32:0) a | C38H77O9P | 0.87 | 1374.76 (687.85) | 458.97 (455.32) |
| 760.5590 | − | M+FA−H [−1] | AS 1-5 a | C40H77NO9 | 1.34 | 1273.41 (530.64) | 489.87 (320.17) |
| 762.5650 | − | M−H [−1] | PS (O−35:0) a | C41H82NO9P | −0.64 | 1541.92 (1105.94) | 674.73 (559.37) |
| 834.5294 | − | M−H [1−] | PS (16:0/16:0) b | C38H74NO10P | −0.41 | 1351.20 (525.54) | 733.77 (538.39) |
| 856.5141 | − | M+Na−2H [−1] | PS (40:6) b | C46H78NO10P | 3.69 | 1575.69 (1472.24) | 463.90 (483.53) |
| 908.6356 | − | M+Na−2H [−1] | PS (43:1) b | C49H94NO10P | −0.63 | 1575.01 (1318.04) | 1016.98 (1203.54) |
| 932.6353 | − | M+FA−H [−1] | PC (44:7) a | C52H90NO8P | −3.75 | 1854.09 (1614.94) | 621.01 (482.13) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mecatti, G.C.; Sánchez-Vinces, S.; Fernandes, A.M.A.P.; Messias, M.C.F.; de Santis, G.K.D.; Porcari, A.M.; Marson, F.A.L.; Carvalho, P.d.O. Potential Lipid Signatures for Diagnosis and Prognosis of Sepsis and Systemic Inflammatory Response Syndrome. Metabolites 2020, 10, 359. https://doi.org/10.3390/metabo10090359
Mecatti GC, Sánchez-Vinces S, Fernandes AMAP, Messias MCF, de Santis GKD, Porcari AM, Marson FAL, Carvalho PdO. Potential Lipid Signatures for Diagnosis and Prognosis of Sepsis and Systemic Inflammatory Response Syndrome. Metabolites. 2020; 10(9):359. https://doi.org/10.3390/metabo10090359
Chicago/Turabian StyleMecatti, Giovana Colozza, Salvador Sánchez-Vinces, Anna Maria A. P. Fernandes, Marcia C. F. Messias, Gabrielle K. D. de Santis, Andreia M. Porcari, Fernando A. L. Marson, and Patrícia de Oliveira Carvalho. 2020. "Potential Lipid Signatures for Diagnosis and Prognosis of Sepsis and Systemic Inflammatory Response Syndrome" Metabolites 10, no. 9: 359. https://doi.org/10.3390/metabo10090359
APA StyleMecatti, G. C., Sánchez-Vinces, S., Fernandes, A. M. A. P., Messias, M. C. F., de Santis, G. K. D., Porcari, A. M., Marson, F. A. L., & Carvalho, P. d. O. (2020). Potential Lipid Signatures for Diagnosis and Prognosis of Sepsis and Systemic Inflammatory Response Syndrome. Metabolites, 10(9), 359. https://doi.org/10.3390/metabo10090359