Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets
Abstract
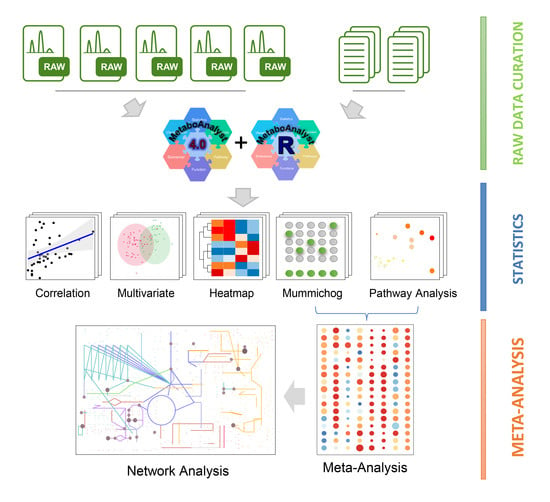
:1. Introduction
2. Results
2.1. Summary of Different Datasets and Their Clinical Characteristics
2.2. Processing and Overview of Individual Datasets
2.3. Metabolic Pathways Changes in COVID-19 Patients
2.4. Identification of Metabolic Hot Spots in COVID-19
2.5. Metabolic Changes between Mild-to-Moderate (MM) and Severe COVID-19
2.6. Exploration of Metabolic Perturbations in Fatal COVID-19
3. Discussion
4. Methods and Materials
4.1. Data Curation
4.2. Patient Classification
4.3. Raw Spectra Processing
4.4. Statistical Analysis
4.5. Metabolic Pathway Analysis and Meta-Analysis
4.6. Global Metabolic Network Visualization
4.7. Cluster Heatmap Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed on 5 November 2020).
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; Veen, W.; Brüggen, M.C.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef]
- Pang, Z.; Wang, G.; Wang, C.; Zhang, W.; Liu, J.; Wang, F. Serum Metabolomics Analysis of Asthma in Different Inflammatory Phenotypes: A Cross-Sectional Study in Northeast China. BioMed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Shu, T.; Yang, X.; Song, J.-X.; Zhang, M.; Yao, C.; Liu, W.; Huang, M.; Yu, Y.; Yang, Q.; et al. Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 2020, 7, 1157–1168. [Google Scholar] [CrossRef]
- Song, J.-W.; Lam, S.M.; Fan, X.; Cao, W.-J.; Wang, S.-Y.; Tian, H.; Chua, G.H.; Zhang, C.; Meng, F.-P.; Xu, Z.; et al. Omics-Driven Systems Interrogation of Metabolic Dysregulation in COVID-19 Pathogenesis. Cell Metab. 2020, 32, 188–202.e5. [Google Scholar] [CrossRef]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef]
- Maras, J.S.; Sharma, S.; Bhat, A.R.; Aggarwal, R.; Gupta, E.; Sarin, S.K. Multi-Omics integration analysis of respiratory specimen characterizes baseline molecular determinants associated with COVID-19 diagnosis. MedRxiv 2020. [Google Scholar] [CrossRef]
- Blasco, H.; Bessy, C.; Plantier, L.; Lefevre, A.; Piver, E.; Bernard, L.; Marlet, J.; Stefic, K.; Benz-De Bretagne, I.; Cannet, P.; et al. The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 2020, 10, 16824. [Google Scholar] [CrossRef]
- Thomas, T.; Stefanoni, D.; Reisz, J.A.; Nemkov, T.; Bertolone, L.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hansen, K.C.; Hod, E.A.; et al. COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 2020, 5, e140327. [Google Scholar] [CrossRef]
- Cai, Y.; Kim, D.J.; Takahashi, T.; Broadhurst, D.I.; Ma, S.; Rattray, N.J.W.; Casanovas-Massana, A.; Israelow, B.; Klein, J.; Lucas, C.; et al. Kynurenic acidunderlies sex-specific immune responsestoCOVID-19. MedRxiv 2020. [Google Scholar] [CrossRef]
- Su, Y.; Chen, D.; Lausted, C.; Yuan, D.; Choi, J.; Dai, C.; Voillet, V.; Scherler, K.; Troisch, P.; Duvvuri, V.R.; et al. Multiomic Immunophenotyping of COVID-19 Patients Reveals Early Infection Trajectories. BioRxiv 2020. [Google Scholar] [CrossRef]
- Zhao, Y.; Shang, Y.; Ren, Y.; Bie, Y.; Qiu, Y.; Yuan, Y.; Zhao, Y.; Zou, L.; Lin, S.H.; Zhou, X. Omics study reveals abnormal alterations of breastmilk proteins and metabolites in puerperant women with COVID-19. Signal Transduct. Target. Ther. 2020, 5, 247. [Google Scholar] [CrossRef]
- Delafiori, J.; Navarro, L.C.; Siciliano, R.F.; de Melo, G.C.; Busanello, E.N.B.; Nicolau, J.C.; Sales, G.M.; de Oliveira, A.N.; Val, F.F.A.; de Oliveira, D.N.; et al. Covid-19 automated diagnosis and risk assessment through Metabolomics and Machine-Learning. MedRxiv 2020. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 2008, 9, 139–150. [Google Scholar] [CrossRef]
- Chiurchiù, V.; Leuti, A.; Maccarrone, M. Bioactive Lipids and Chronic Inflammation: Managing the Fire Within. Front. Immunol. 2018, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Struwe, W.; Emmott, E.; Bailey, M.; Sharon, M.; Sinz, A.; Corrales, F.J.; Thalassinos, K.; Braybrook, J.; Mills, C.; Barran, P.; et al. The COVID-19 MS Coalition-accelerating diagnostics, prognostics, and treatment. Lancet 2020, 395, 1761–1762. [Google Scholar] [CrossRef]
- Zaas, A.K.; Chen, M.; Varkey, J.; Veldman, T.; Hero, A.O., 3rd; Lucas, J.; Huang, Y.; Turner, R.; Gilbert, A.; Lambkin-Williams, R.; et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 2009, 6, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Zhou, G.; Stevenson, M.M.; Geary, T.G.; Xia, J. Comprehensive Transcriptome Meta-analysis to Characterize Host Immune Responses in Helminth Infections. PLoS Negl. Trop. Dis. 2016, 10, e0004624. [Google Scholar] [CrossRef]
- Gardinassi, L.G.; Souza, C.O.S.; Sales-Campos, H.; Fonseca, S.G. Immune and Metabolic Signatures of COVID-19 Revealed by Transcriptomics Data Reuse. Front. Immunol. 2020, 11, 1636. [Google Scholar] [CrossRef]
- Cavalli, E.; Petralia, M.C.; Basile, M.S.; Bramanti, A.; Bramanti, P.; Nicoletti, F.; Spandidos, D.A.; Shoenfeld, Y.; Fagone, P. Transcriptomic analysis of COVID19 lungs and bronchoalveolar lavage fluid samples reveals predominant B cell activation responses to infection. Int. J. Mol. Med. 2020, 46, 1266–1273. [Google Scholar] [CrossRef]
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Peron, J.P.S.; Nakaya, H.I. ACE2 Expression is Increased in the Lungs of Patients with Comorbidities Associated with Severe COVID-19. MedRxiv Prepr. Serv. Health Sci. 2020. [Google Scholar] [CrossRef]
- Kale, N.S.; Haug, K.; Conesa, P.; Jayseelan, K.; Moreno, P.; Rocca-Serra, P.; Nainala, V.C.; Spicer, R.A.; Williams, M.; Li, X.; et al. MetaboLights: An Open-Access Database Repository for Metabolomics Data. Curr. Protoc. Bioinform. 2016, 53. [Google Scholar] [CrossRef]
- Gauglitz, J.M.; Bittremieux, W.; Williams, C.L.; Weldon, K.C.; Panitchpakdi, M.; Di Ottavio, F.; Aceves, C.M.; Brown, E.; Sikora, N.C.; Jarmusch, A.K.; et al. Reference data based insights expand understanding of human metabolomes. BioRxiv 2020. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef] [Green Version]
- Yu, T.; Park, Y.; Johnson, J.M.; Jones, D.P. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics 2009, 25, 1930–1936. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [Green Version]
- García, L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020, 11, 1441. [Google Scholar] [CrossRef]
- San Juan, I.; Bruzzone, C.; Bizkarguenaga, M.; Bernardo-Seisdedos, G.; Laín, A.; Gil-Redondo, R.; Diercks, T.; Gil-Martínez, J.; Urquiza, P.; Arana, E.; et al. Abnormal concentration of porphyrins in serum from COVID-19 patients. Br. J. Haematol. 2020, 190, e265–e267. [Google Scholar] [CrossRef]
- Ponti, G.; Maccaferri, M.; Ruini, C.; Tomasi, A.; Ozben, T. Biomarkers associated with COVID-19 disease progression. Crit Rev. Clin. Lab. Sci 2020, 57, 389–399. [Google Scholar] [CrossRef]
- liu, w.; Li, h. COVID-19: Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism. ChemRxiv 2020. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Hoxha, M. What about COVID-19 and arachidonic acid pathway? Eur. J. Clin. Pharmacol. 2020, 76, 1501–1504. [Google Scholar] [CrossRef] [PubMed]
- Hammock, B.D.; Wang, W.; Gilligan, M.M.; Panigrahy, D. Eicosanoids: The Overlooked Storm in Coronavirus Disease 2019 (COVID-19)? Am. J. Pathol. 2020, 190, 1782–1788. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.; Sharma, L.; Roberts, L.; Peng, X.; Bermejo, S.; Leighton, I.; Massana, A.C.; Farhadian, S.; Ko, A.; DelaCruz, C.; et al. Severe SARS-CoV-2 infection in humans is defined by a shift in the serum lipidome resulting in dysregulation of eicosanoid immune mediators. MedRxiv 2020. [Google Scholar] [CrossRef]
- Darwesh, A.M.; Bassiouni, W.; Sosnowski, D.K.; Seubert, J.M. Can N-3 polyunsaturated fatty acids be considered a potential adjuvant therapy for COVID-19-associated cardiovascular complications? Pharm. Ther. 2020, 107703. [Google Scholar] [CrossRef] [PubMed]
- Margină, D.; Ungurianu, A.; Purdel, C.; Tsoukalas, D.; Sarandi, E.; Thanasoula, M.; Tekos, F.; Mesnage, R.; Kouretas, D.; Tsatsakis, A. Chronic Inflammation in the Context of Everyday Life: Dietary Changes as Mitigating Factors. Int. J. Environ. Res. Public Health 2020, 17, 4135. [Google Scholar] [CrossRef]
- Iddir, M.; Brito, A.; Dingeo, G.; Fernandez Del Campo, S.S.; Samouda, H.; La Frano, M.R.; Bohn, T. Strengthening the Immune System and Reducing Inflammation and Oxidative Stress through Diet and Nutrition: Considerations during the COVID-19 Crisis. Nutrients 2020, 12, 1562. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19-possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef]
- Gassen, N.C.; Papies, J.; Bajaj, T.; Dethloff, F.; Emanuel, J.; Weckmann, K.; Heinz, D.E.; Heinemann, N.; Lennarz, M.; Richter, A.; et al. Analysis of SARS-CoV-2-controlled autophagy reveals spermidine, MK-2206, and niclosamide as putative antiviral therapeutics. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Liu, G.; Wang, L.; Liang, Y.; Zhou, Q.; Wu, F.; Yao, J.; Chen, B. From the insight of glucose metabolism disorder: Oxygen therapy and blood glucose monitoring are crucial for quarantined COVID-19 patients. Ecotoxicol. Environ. Saf. 2020, 197, 110614. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.C.; O’Neill, L.A.J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Suzuki, N.; Ogra, Y. Effect of gut microflora on nutritional availability of selenium. Food Chem. 2020, 319, 126537. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Chander, B. COVID 19 mortality: Probable role of microbiome to explain disparity. Med. Hypotheses 2020, 144, 110209. [Google Scholar] [CrossRef] [PubMed]
- Bradley, B.T.; Maioli, H.; Johnston, R.; Chaudhry, I.; Fink, S.L.; Xu, H.; Najafian, B.; Deutsch, G.; Lacy, J.M.; Williams, T.; et al. Histopathology and ultrastructural findings of fatal COVID-19 infections in Washington State: A case series. Lancet 2020, 396, 320–332. [Google Scholar] [CrossRef]
- Polonikov, A. Endogenous Deficiency of Glutathione as the Most Likely Cause of Serious Manifestations and Death in COVID-19 Patients. ACS Infect. Dis. 2020, 6, 1558–1562. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Abdulrab, S.; Al-Maweri, S.; Halboub, E. Ursodeoxycholic acid as a candidate therapeutic to alleviate and/or prevent COVID-19-associated cytokine storm. Med. Hypotheses 2020, 143, 109897. [Google Scholar] [CrossRef]
- Ouyang, L.; Gong, J. Mitochondrial-targeted ubiquinone: A potential treatment for COVID-19. Med. Hypotheses 2020, 144, 110161. [Google Scholar] [CrossRef]
- Kumrungsee, T.; Zhang, P.; Chartkul, M.; Yanaka, N.; Kato, N. Potential Role of Vitamin B6 in Ameliorating the Severity of COVID-19 and Its Complications. Front. Nutr. 2020, 7, 562051. [Google Scholar] [CrossRef]
- Thair, S.A.; He, Y.D.; Hasin-Brumshtein, Y.; Sakaram, S.; Pandya, R.; Toh, J.; Rawling, D.; Remmel, M.; Coyle, S.; Dalekos, G.N.; et al. Transcriptomic similarities and differences in host response between SARS-CoV-2 and other viral infections. iScience 2021, 24, 101947. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The, P.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Health Commission of the People’s Republic of China. Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (8th). Available online: http://www.nhc.gov.cn/ (accessed on 5 November 2020).
- WHO. WHO R&D Blueprint novel Coronavirus COVID-19 Therapeutic Trial Synopsis. Available online: https://www.who.int/blueprint/priority-diseases/key-action/COVID-19_Treatment_Trial_Design_Master_Protocol_synopsis_Final_18022020.pdf (accessed on 5 November 2020).
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [Green Version]






| Datasets | Chromatogram | MS | Patient Classification | Country | ||||
|---|---|---|---|---|---|---|---|---|
| Total | HC | MM | Severe | Fatal | ||||
| A1 [12] | UPLC-C18 | Q/E | 49 | 16 | 27 | 6 | 0 | USA |
| A2 * [13] | UPLC-HILIC | Q/TOF | 59 | 20 | 39 | 0 | 0 | USA |
| A3 * [13] | UPLC-C18 | |||||||
| B1 [26] | HPLC- C18 | micrOTOF | 28 | 13 | 6 | 3 | 6 | Brazil |
| C1 [8] | UPLC-C18 | Triple TOF | 76 | 26 | 37 | 11 | 2 | China |
| C2 ** [9] | UPLC-C18 | QE-HF | 71 | 25 | 37 | 28 | 0 | China |
| C3 ** [7] | UPLC- C30 | Q/TRAP | 96 | 10 | 14 | 11 | 9 | China |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pang, Z.; Zhou, G.; Chong, J.; Xia, J. Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets. Metabolites 2021, 11, 44. https://doi.org/10.3390/metabo11010044
Pang Z, Zhou G, Chong J, Xia J. Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets. Metabolites. 2021; 11(1):44. https://doi.org/10.3390/metabo11010044
Chicago/Turabian StylePang, Zhiqiang, Guangyan Zhou, Jasmine Chong, and Jianguo Xia. 2021. "Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets" Metabolites 11, no. 1: 44. https://doi.org/10.3390/metabo11010044
APA StylePang, Z., Zhou, G., Chong, J., & Xia, J. (2021). Comprehensive Meta-Analysis of COVID-19 Global Metabolomics Datasets. Metabolites, 11(1), 44. https://doi.org/10.3390/metabo11010044