Does DDI-Predictor Help Pharmacists to Detect Drug-Drug Interactions and Resolve Medication Issues More Effectively?
Abstract
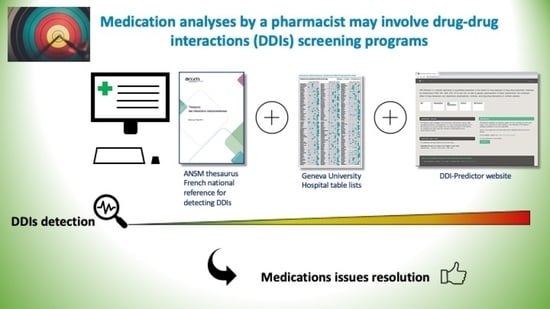
:1. Introduction
2. Results
2.1. Description of the Interactions
2.2. Description of the PIs and PAR Associated with DDI-Predictor
3. Discussion
4. Materials and Methods
4.1. The DDI Process Screening
4.2. Description of the Tools Used
4.3. Data Collection
4.4. Data Coding
4.5. Data Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pirmohamed, M.; James, S.; Meakin, S.; Green, C.; Scott, A.K.; Walley, T.J.; Farrar, K.; Park, B.K.; Breckenridge, A.M. Adverse drug reactions as cause of admission to hospital: Prospective analysis of 18 820 patients. BMJ 2004, 329, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Michel, P.; Lathelize, M.; Domecq, S.; Kret, M.; Bru-Sonnet, R.; Quenon, J.-L.; Minodier, C.; Moty-Monnereau, C.; Chaleix, M.; Olier, L.; et al. «Les Evénements Indésirables Graves dans les Etablissements de Santé: Fréquence, Evitabilité et Acceptabilité», Études et Résultats, DREES, n° 761, mai. 2011. Available online: http://www.epsilon.insee.fr/jspui/bitstream/1/13389/1/article201017.pdf (accessed on 16 December 2020).
- Michel, P.; Minodier, C.; Lathelize, M.; Moty-Monnereau, C.; Domecq, S.; Chaleix, M.; Izotte-kret, M.; Bru-Sonnet, R.; Quenon, J.-L.; Olier, L. «Les Evénements Indésirables Graves Associés aux soins Observés dans les Etablissements de Santé», Dossiers Solidarité et Santé, DREES, n° 17. 2010. Available online: http://www.epsilon.insee.fr/jspui/bitstream/1/13389/1/article201017.pdf (accessed on 16 December 2020).
- Strandell, J.; Wahlin, S. Pharmacodynamic and pharmacokinetic drug interactions reported to VigiBase, the WHO global individual case safety report database. Eur. J. Clin. Pharmacol. 2011, 67, 633–641. [Google Scholar] [CrossRef] [Green Version]
- Rendic, S. Summary of information on human CYP enzymes: Human P450 metabolism data. Drug Metab. Rev. 2002, 34, 83–448. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Oscarson, M.; McLellan, R.A. Polymorphic human cytochrome P450 enzymes: An opportunity for individualized drug treatment. Trends Pharmacol. Sci. 1999, 20, 342–349. [Google Scholar] [CrossRef]
- Liu, R.; AbdulHameed, M.D.M.; Kumar, K.; Yu, X.; Wallqvist, A.; Reifman, J. Data-driven prediction of adverse drug reactions induced by drug-drug interactions. BMC Pharmacol. Toxicol. 2017, 18, 44. [Google Scholar] [CrossRef] [Green Version]
- Hohl, C.M.; Dankoff, J.; Colacone, A.; Afilalo, M. Polypharmacy, adverse drug-related events, and potential adverse drug interactions in elderly patients presenting to an emergency department. Ann. Emerg. Med. 2001, 38, 666–671. [Google Scholar] [CrossRef]
- De Leon, J.; Barnhill, J.; Rogers, T.; Boyle, J.; Chou, W.H.; Wedlund, P.J. Pilot study of the cytochrome P450-2D6 genotype in a psychiatric state hospital. Am. J. Psychiatry 1998, 155, 1278–1280. [Google Scholar] [CrossRef]
- Ozdil, B.; Akkiz, H.; Bayram, S.; Bekar, A.; Akgöllü, E.; Sandikçi, M. Influence of CYP2C19 functional polymorphism on Helicobacter pylori eradication. Turk J. Gastroenterol. 2010, 21, 23–28. [Google Scholar] [CrossRef]
- Franz, C.C.; Egger, S.; Born, C.; Rätz Bravo, A.E.; Krähenbühl, S. Potential drug-drug interactions and adverse drug reactions in patients with liver cirrhosis. Eur. J. Clin. Pharmacol. 2012, 68, 179–188. [Google Scholar] [CrossRef] [Green Version]
- Robiek, T.; Vaupotic, T.; Mrahr, A.; Lainscak, M. Drug-drug interaction software in clinical practice: A systematic review. Eur. J. Clin. Pharmacol. 2015, 71, 131–142. [Google Scholar]
- Zakrzewski-Jakubiak, H.; Doan, J.; Lamoureux, P.; Singh, D.; Turgeon, J.; Tannenbaum, C. Detection and prevention of drug-drug interactions in the hospitalized elderly: Utility of new cytochrome p450-based software. Am. J. Geriatr. Pharmacother. 2011, 9, 461–470. [Google Scholar] [CrossRef]
- Beeler, P.A.; Eschmann, E.; Rosen, C.; Blaser, J. Use of an on-demand drug-drug interaction checker by prescribers and consultants: A retrospective analysis in a Swiss teaching hospital. Drug Saf. 2013, 36, 427–434. [Google Scholar] [CrossRef] [Green Version]
- Thésaurus des Interactions Médicamenteuses de l’ANSM. Available online: http://ansm.sante.fr/var/ansm_site/storage/original/application/de444ea9eb4bc084905c917c902a805f.pdf (accessed on 23 June 2020).
- Hôpitaux Universitaires de Genève. Interactions Médicamenteuses, Cytochromes P450 et P-Glycoprotéine (Pgp). Available online: https://www.hug.ch/sites/interhug/files/structures/pharmacologie_et_toxicologie_cliniques/a5_cytochromes_6_2.pdf (accessed on 23 June 2020).
- Tod, M.; Pierrillas, P.B.; Bourguignon, L.; Goutelle, S. Comparison of the static in vivo approach to a physiologically based pharmacokinetic approach for metabolic drug–drug interactions prediction. Int. J. Pharm. 2016, 1, 25–34. [Google Scholar]
- Loue, C.; Tod, M. Reliability and extension of quantitative prediction of CYP3A4-mediated drug interactions based on clinical data. AAPS J. 2014, 16, 1309–1320. [Google Scholar] [CrossRef] [Green Version]
- Steelandt, J.; Jean-Bart, E.; Goutelle, S.; Tod, M. A Prediction Model of Drug Exposure in Cirrhotic Patients According to Child-Pugh Classification. Clin. Pharm. 2015, 54, 1245–1258. [Google Scholar] [CrossRef]
- Bond, C.A.; Raehl, C.L. Clinical pharmacy services, pharmacy staffing, and hospital mortality rates. Pharmacotherapy 2007, 27, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Egger, S.S.; Drewe, J.; Schlienger, R.G. Potential drug-drug interactions in the medication of medical patients at hospital discharge. Eur. J. Clin. Pharmacol. 2003, 58, 773–778. [Google Scholar] [CrossRef]
- Cornu, P.; Steurbaut, S.; Soštarić, S.; Mrhar, A.; Dupont, A.G. Performance of a clinical decision support system and of clinical pharmacists in preventing drug-drug interactions on a geriatric ward. Int. J. Clin. Pharm. 2014, 36, 519–525. [Google Scholar] [CrossRef]
- Olakotan, O.; Mohd Yusof, M.; Ezat Wan Puteh, S. A Systematic Review on CDSS Alert Appropriateness. Stud. Health Technol. Inform. 2020, 270, 906–910. [Google Scholar]
- Sola Bonada, N.; Álvarez Díaz, A.M.; Codina Jané, C. The role of the Pharmacist in the design, development and implementation of Medication Prescription Support Systems. Farm Hosp. 2016, 40, 457–476. [Google Scholar]
- Michalets, E.L. Update: Clinically significant cytochrome P-450 drug interactions. Pharmacotherapy 1998, 18, 84–112. [Google Scholar]
- Baciewicz, A.M.; Chrisman, C.R.; Finch, C.K.; Self, T.H. Update on rifampin, rifabutin, and rifapentine drug interactions. Curr. Med. Res. Opin. 2013, 29, 1–12. [Google Scholar] [CrossRef]
- Riesenman, C. Antidepressant drug interactions and the cytochrome P450 system: A critical appraisal. Pharmacotherapy 1995, 15, 84S–99S. [Google Scholar]
- Müller, J.; Keiser, M.; Drozdzik, M.; Oswald, S. Expression, regulation and function of intestinal drug transporters: An update. Biol. Chem. 2017, 398, 175–192. [Google Scholar] [CrossRef] [PubMed]
- Andes, D.; Marchillo, K.; Stamstad, T.; Conklin, R. In vivo pharmacokinetics and pharmacodynamics of a new triazole, voriconazole, in a murine candidiasis model. Antimicrob Agents Chemother 2003, 47, 3165–3169. [Google Scholar] [CrossRef] [Green Version]
- Tod, M.; Nkoud-Mongo, C.; Gueyffier, F. Impact of genetic polymorphism on drug-drug interactions mediated by cytochromes: A general approach. AAPS J. 2013, 15, 1242–1252. [Google Scholar] [CrossRef] [Green Version]
- Tod, M.; Bourguignon, L.; Bleyzac, N.; Goutelle, S. A model for predicting the interindividual variability of drug-drug interactions. AAPS J. 2017, 19, 497–509. [Google Scholar] [CrossRef]


| ANSM | GUH | DDI-P | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| - | N | 1 | 2 | 3 | 4 | ND | NR | N | RAUC ≤ 0.5 | RAUC 0.5–2 | RAUC > 2 | NR |
| 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | - | - | - |
| 4 | 2 | 0 | 0 | 0 | 0 | 2 | 4 | - | - | 3.44 ± 1.15 | 0 | |
| 2 | 12 | 8 | 3 | 0 | 0 | 0 | 1 | 10 | 0.22 ± 0.06 | 1 | - | 2 |
| 10 | 9 | 0 | 0 | 0 | 0 | 1 | 9 | 1.74 | 6.10 ± 5.47 | 1 | ||
| 3 | 18 | 9 | 1 | 4 | 0 | 2 | 2 | 15 | 0.31 ± 0.06 | 0.69 ± 0.12 | - | 3 |
| 26 | 18 | 2 | 2 | 0 | 0 | 4 | 21 | - | 1.71 ± 0.38 | 5.03 ± 1.97 | 5 | |
| 4 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0.5 | - | - | 1 |
| 20 | 5 | 8 | 4 | 2 | 0 | 1 | 19 | - | 1.47 ± 0.1 | 2.56 ± 0.84 | 1 | |
| ND | 35 | 15 | 2 | 0 | 1 | 5 | 12 | 29 | 0.29 ± 0.04 | 0.765 ± 0.24 | - | 6 |
| 100 | 44 | 22 | 12 | 3 | 10 | 9 | 85 | - | 1.46 ± 0.07 | 4.17 ± 1.14 | 15 | |
| NR | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | - | - | - | 0 |
| 3 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | - | 1.19 ± 1.97 | 6.13 | 0 | |
| RAUC ≤ 0.5 | RAUC 0.5–2 | RAUC > 2 | ||||
|---|---|---|---|---|---|---|
| Median | 0.28 | 1.51 | 3.05 | |||
| N | % | N | % | N | % | |
| PIs | 36/40 | 90.0 † | 46/104 | 44.2 † | 39/52 | 75.0 † |
| Dose increase | 8/9 | 88.9 | 1/1 | 100 | 0/0 | - |
| Change in the substrate | 13/15 | 86.7 | 0/0 | - | 7/7 | 100 |
| Dose decrease | 0/0 | - | 4/6 | 66.7 | 11/12 | 91.7 |
| Less frequent dosing | 1/1 | 100 | 0/0 | - | 0/0 | - |
| Change in the interactor | 0/4 | 2/2 | 100 | 3/3 | 100 | |
| Therapeutic drug monitoring | 4/4 | 100 | 8/9 | 88.9 | 2/4 | 50 |
| Adverse drug reaction monitoring | 5/5 | 100 | 24/28 | 86 | 9/13 | 69 |
| Withdrawal of the interactor | 1/2 | 50 | - | - | - | - |
| PAR | 32/36 | 88.8 ‡ | 39/46 | 84.8 ‡ | 32/39 | 82.0 ‡ |
| RAUC ≤ 0.5 (N = 36) | RAUC 0.5–2 (N = 46) | RAUC > 2 (N = 39) | Total | ||||
|---|---|---|---|---|---|---|---|
| Detected | PIs | PAR | PIs | PAR | PIs | PAR | - |
| ANSM | 13 a (36.1%) | 13/13 | 17 b (37.0%) | 15/17 | 20 c (51.3%) | 13/20 | 50 (41.3%) |
| GUH | 25 a (69.4%) | 21/25 | 37 b (80.4%) | 31/37 | 36 c (92.3%) | 30/36 | 98 (81.0%) |
| Not detected | - | - | - | - | - | - | - |
| ANSM | 23 (63.9%) | 19/23 | 29 (63.0%) | 24/29 | 19 (48.7%) | 17/19 | 71 (58.7%) |
| GUH | 11 (30.6%) | 11/11 | 9 (19.6%) | 8/9 | 3 (7.7%) | 2/3 | 23 (19.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreau, F.; Simon, N.; Walther, J.; Dambrine, M.; Kosmalski, G.; Genay, S.; Perez, M.; Lecoutre, D.; Belaiche, S.; Rousselière, C.; et al. Does DDI-Predictor Help Pharmacists to Detect Drug-Drug Interactions and Resolve Medication Issues More Effectively? Metabolites 2021, 11, 173. https://doi.org/10.3390/metabo11030173
Moreau F, Simon N, Walther J, Dambrine M, Kosmalski G, Genay S, Perez M, Lecoutre D, Belaiche S, Rousselière C, et al. Does DDI-Predictor Help Pharmacists to Detect Drug-Drug Interactions and Resolve Medication Issues More Effectively? Metabolites. 2021; 11(3):173. https://doi.org/10.3390/metabo11030173
Chicago/Turabian StyleMoreau, Fanny, Nicolas Simon, Julia Walther, Mathilde Dambrine, Gaetan Kosmalski, Stéphanie Genay, Maxime Perez, Dominique Lecoutre, Stéphanie Belaiche, Chloé Rousselière, and et al. 2021. "Does DDI-Predictor Help Pharmacists to Detect Drug-Drug Interactions and Resolve Medication Issues More Effectively?" Metabolites 11, no. 3: 173. https://doi.org/10.3390/metabo11030173
APA StyleMoreau, F., Simon, N., Walther, J., Dambrine, M., Kosmalski, G., Genay, S., Perez, M., Lecoutre, D., Belaiche, S., Rousselière, C., Tod, M., Décaudin, B., & Odou, P. (2021). Does DDI-Predictor Help Pharmacists to Detect Drug-Drug Interactions and Resolve Medication Issues More Effectively? Metabolites, 11(3), 173. https://doi.org/10.3390/metabo11030173