Nontargeted Metabolomics as a Screening Tool for Estimating Bioactive Metabolites in the Extracts of 50 Indigenous Korean Plants
Abstract
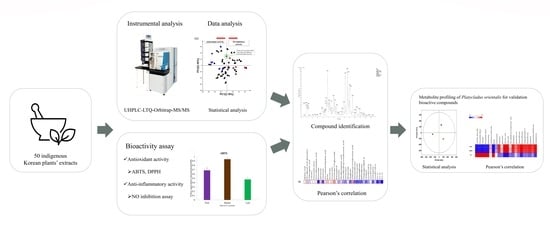
:1. Introduction
2. Results and Discussion
2.1. Bioactivities of Indigenous Korean Plant Species
2.2. Metabolite Profiling of the Bioactivities of 50 Indigenous Korean Plants
2.3. Bioactivity Correlations for the Secondary Metabolites of Indigenous Korean Plants
2.4. Bioactivity Correlations with the Secondary Metabolties of P. orientalis for the Validation of Bioactive Compounds in Indigenous Korean Plants
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Plant Materials
3.3. UHPLC-LTQ-Orbitrap-MS/MS Analysis
3.4. Data Processing and Statistical Analysis
3.5. Antioxidant Assays
3.6. Nitric Oxide (NO) Inhibition Assays
3.7. MTT Assays for Measuring Cell Viability
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Park, K.-Y.; Jeong, J.-K.; Lee, Y.-E.; Daily, J.W., III. Health benefits of kimchi (Korean fermented vegetables) as a probiotic food. J. Med. Food 2014, 17, 6–20. [Google Scholar] [CrossRef]
- Choi, C.W.; Kim, S.C.; Hwang, S.S.; Choi, B.K.; Ahn, H.J.; Lee, M.Y.; Park, S.H.; Kim, S.K. Antioxidant activity and free radical scavenging capacity between Korean medicinal plants and flavonoids by assay-guided comparison. Plant Sci. 2002, 163, 1161–1168. [Google Scholar] [CrossRef]
- Lee, K.; Kim, B.; Kim, J.; Heo, M.; Kim, H. Biological screening of 100 plant extracts for cosmetic use (I): Inhibitory activities of tyrosinase and DOPA auto-oxidation. Int. J. Cosmet. Sci. 1997, 19, 291–298. [Google Scholar] [CrossRef]
- Cowan, M.M. Plant products as antimicrobial agents. Clin. Microbiol. Rev. 1999, 12, 564–582. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Mumper, R.J. Plant phenolics: Extraction, analysis and their antioxidant and anticancer properties. Molecules 2010, 15, 7313–7352. [Google Scholar] [CrossRef]
- Korkina, L.; Kostyuk, V.; De Luca, C.; Pastore, S. Plant phenylpropanoids as emerging anti-inflammatory agents. Mini Rev. Med. Chem. 2011, 11, 823–835. [Google Scholar] [CrossRef] [PubMed]
- Seca, A.M.; Pinto, D.C. Plant secondary metabolites as anticancer agents: Successes in clinical trials and therapeutic application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tungmunnithum, D.; Pinthong, D.; Hano, C. Flavonoids from Nelumbo nucifera Gaertn., a medicinal plant: Uses in traditional medicine, phytochemistry and pharmacological activities. Medicines 2018, 5, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kallscheuer, N.; Classen, T.; Drepper, T.; Marienhagen, J. Production of plant metabolites with applications in the food industry using engineered microorganisms. Curr. Opin. Biotechnol. 2019, 56, 7–17. [Google Scholar] [CrossRef]
- Zhou, Y.X.; Zhang, H.; Peng, C. Puerarin: A review of pharmacological effects. Phytother. Res. 2014, 28, 961–975. [Google Scholar] [CrossRef]
- Takshak, S.; Agrawal, S. Defense potential of secondary metabolites in medicinal plants under UV-B stress. J. Photochem. Photobiol. B Biol. 2019, 193, 51–88. [Google Scholar] [CrossRef]
- Nijveldt, R.J.; Van Nood, E.; Van Hoorn, D.E.; Boelens, P.G.; Van Norren, K.; Van Leeuwen, P.A. Flavonoids: A review of probable mechanisms of action and potential applications. Am. J. Clin. Nutr. 2001, 74, 418–425. [Google Scholar] [CrossRef]
- Stuehr, D.; Nathan, C. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J. Exp. Med. 1989, 169, 1543–1555. [Google Scholar] [CrossRef]
- Nathan, C.F.; Hibbs, J.B., Jr. Role of nitric oxide synthesis in macrophage antimicrobial activity. Curr. Opin. Immunol. 1991, 3, 65–70. [Google Scholar] [CrossRef]
- MacMicking, J.; Xie, Q.-W.; Nathan, C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997, 15, 323–350. [Google Scholar] [CrossRef]
- Seifried, H.E.; Anderson, D.E.; Fisher, E.I.; Milner, J.A. A review of the interaction among dietary antioxidants and reactive oxygen species. J. Nutr. Biochem. 2007, 18, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Padzil, A.; Syahida, A.; Abdullah, N.; Zuhainis, S.; Maziah, M.; Sulaiman, M.; Israf, D.; Shaari, K.; Lajis, N. Evaluation of anti-inflammatory, antioxidant and anti-nociceptive activities of six Malaysian medicinal plants. J. Med. Plants Res. 2011, 5, 5555–5563. [Google Scholar]
- Yanishlieva, N.V.; Marinova, E.; Pokorný, J. Natural antioxidants from herbs and spices. Eur. J. Lipid Sci. Technol. 2006, 108, 776–793. [Google Scholar] [CrossRef]
- Akhtar, N.; Mirza, B. Phytochemical analysis and comprehensive evaluation of antimicrobial and antioxidant properties of 61 medicinal plant species. Arab. J. Chem. 2018, 11, 1223–1235. [Google Scholar] [CrossRef] [Green Version]
- Sulaiman, M.; Tijani, H.I.; Abubakar, B.M.; Haruna, S.; Hindatu, Y.; Mohammed, J.N.; Idris, A. An overview of natural plant antioxidants: Analysis and evaluation. Adv. Biochem. 2013, 1, 64–72. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.H.; Hur, S.K.; Oh, O.-J.; Kim, S.S.; Nam, K.A.; Lee, S.K. Evaluation of natural products on inhibition of inducible cyclooxygenase (COX-2) and nitric oxide synthase (iNOS) in cultured mouse macrophage cells. J. Ethnopharmacol. 2002, 83, 153–159. [Google Scholar] [CrossRef]
- Kim, H.-J.; Lee, D.-J.; Ku, J.-J.; Choi, K.; Park, K.-W.; Kang, S.-H.; Moon, C.; Lee, P.-J. Anti-inflammatory effect of extracts from folk plants in Ulleung island. Korean J. Plant Resour. 2013, 26, 169–177. [Google Scholar] [CrossRef] [Green Version]
- Yang, E.-J.; Yim, E.-Y.; Song, G.; Kim, G.-O.; Hyun, C.-G. Inhibition of nitric oxide production in lipopolysaccharide-activated RAW 264.7 macrophages by Jeju plant extracts. Interdiscip. Toxicol. 2009, 2, 245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnaiah, D.; Sarbatly, R.; Nithyanandam, R. A review of the antioxidant potential of medicinal plant species. Food Bioprod. Process. 2011, 89, 217–233. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.-G.; Singh, D.; Lee, J.S.; Lee, S.; Lee, C.H. Exploring the metabolomic diversity of plant species across spatial (leaf and stem) components and phylogenic groups. BMC Plant Biol. 2020, 20, 39. [Google Scholar] [CrossRef] [PubMed]
- Abd Ghafar, S.Z.; Mediani, A.; Ramli, N.S.; Abas, F. Antioxidant, α-glucosidase, and nitric oxide inhibitory activities of Phyllanthus acidus and LC–MS/MS profile of the active extract. Food Biosci. 2018, 25, 134–140. [Google Scholar] [CrossRef]
- Yoo, T.-K.; Jeong, W.T.; Kim, J.G.; Ji, H.S.; Ahn, M.-A.; Chung, J.-W.; Lim, H.B.; Hyun, T.K. UPLC-ESI-Q-TOF-MS-Based Metabolite Profiling, Antioxidant and Anti-Inflammatory Properties of Different Organ Extracts of Abeliophyllum distichum. Antioxidants 2021, 10, 70. [Google Scholar] [CrossRef]
- Yuk, H.J.; Oh, K.Y.; Kim, D.-Y.; Song, H.-H.; Kim, J.Y.; Oh, S.-R.; Ryu, H.W. Metabolomic profiling, antioxidant and anti-inflammatory activities of Hypericum species growing in South Korea. Nat. Prod. Commun. 2017, 12, 1934578X1701200710. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Oh, D.-G.; Lee, S.; Kim, G.R.; Lee, J.S.; Son, Y.K.; Bae, C.-H.; Yeo, J.; Lee, C.H. Chemotaxonomic metabolite profiling of 62 indigenous plant species and its correlation with bioactivities. Molecules 2015, 20, 19719–19734. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.M.; Lee, J.; Park, S.-H.; Lee, C.; Lee, J.W.; Lee, D.; Kim, N.; Lee, D.; Kim, H.Y.; Lee, C.H. LC–MS-based chemotaxonomic classification of wild-type Lespedeza sp. and its correlation with genotype. Plant Cell Rep. 2012, 31, 2085–2097. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.-G.; Singh, D.; Lee, H.J.; Kim, G.R.; Lee, S.; Lee, J.S.; Lee, C.H. Untargeted Metabolomics Toward Systematic Characterization of Antioxidant Compounds in Betulaceae Family Plant Extracts. Metabolites 2019, 9, 186. [Google Scholar] [CrossRef] [Green Version]
- Son, S.Y.; Kim, N.K.; Lee, S.; Singh, D.; Kim, G.R.; Lee, J.S.; Yang, H.-S.; Yeo, J.; Lee, S.; Lee, C.H. Metabolite fingerprinting, pathway analyses, and bioactivity correlations for plant species belonging to the Cornaceae, Fabaceae, and Rosaceae families. Plant Cell Rep. 2016, 35, 1917–1931. [Google Scholar] [CrossRef] [PubMed]
- Won, J.Y.; Son, S.Y.; Lee, S.; Singh, D.; Lee, S.; Lee, J.S.; Lee, C.H. Strategy for screening of antioxidant compounds from two Ulmaceae species based on liquid chromatography-mass spectrometry. Molecules 2018, 23, 1830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, C.; Khatana, S.; Vijayvergia, R. Bioactivity of secondary metabolites of various plants: A review. Int. J. Pharm. Sci. Res. 2019, 10, 494–498. [Google Scholar]
- Bing, F.-H.; Liu, J.; Li, Z.; Zhang, G.-B.; Liao, Y.-F.; Li, J.; Dong, C.-Y. Anti-influenza-virus activity of total alkaloids from Commelina communis L. Arch. Virol. 2009, 154, 1837–1840. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.-Y.; Zeng, H.-W.; Pei, Y.-H.; Li, L.; Ye, J.; Pan, Y.-X.; Zhang, J.-G.; Yuan, X.; Zhang, W.-D. The anti-inflammatory activities of an extract and compounds isolated from Platycladus orientalis (Linnaeus) Franco in vitro and ex vivo. J. Ethnopharmacol. 2012, 141, 647–652. [Google Scholar] [CrossRef]
- He, X.; Fang, J.; Huang, L.; Wang, J.; Huang, X. Sophora flavescens Ait.: Traditional usage, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2015, 172, 10–29. [Google Scholar] [CrossRef]
- Jin, K.-S.; Oh, Y.N.; Park, J.; Lee, J.Y.; Jin, S.; Hyun, S.K.; Hwang, H.J.; Kwon, H.J.; Kim, B.W. Anti-oxidant, anti-melanogenic, and anti-inflammatory activities of Zanthoxylum schinifolium extract and its solvent fractions. Microbiol. Biotechnol. Lett. 2012, 40, 371–379. [Google Scholar] [CrossRef]
- Kim, M.J.; Wang, H.S.; Lee, M.W. Anti-inflammatory effects of fermented bark of Acanthopanax sessiliflorus and its isolated compounds on lipopolysaccharide-treated RAW 264.7 macrophage cells. Evid.-Based Complementary Altern. Med. 2020, 2020, 674925. [Google Scholar] [CrossRef]
- Venkatesan, T.; Choi, Y.-W.; Lee, J.; Kim, Y.-K. Pinus densiflora needle supercritical fluid extract suppresses the expression of pro-inflammatory mediators iNOS, IL-6 and IL-1β, and activation of inflammatory STAT1 and STAT3 signaling proteins in bacterial lipopolysaccharide-challenged murine macrophages. Daru J. Pharm. Sci. 2017, 25, 1–10. [Google Scholar]
- Ozen, T.; Yenigun, S.; Altun, M.; Demirtas, I. Phytochemical Constituents, ChEs and Urease Inhibitions, Antiproliferative and Antioxidant Properties of Elaeagnus umbellata Thunb. Comb. Chem. High Throughput Screen. 2017, 20, 559–578. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Liao, L.; Shang, S.; Zheng, Y.; Sha, W.; Yuan, E. Purification, characterization, and bioactivities of polyphenols from Platycladus orientalis (L.) Franco. J. Food Sci. 2019, 84, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Tuyen, P.T.; Xuan, T.D.; Khang, D.T.; Ahmad, A.; Quan, N.V.; Tu Anh, T.T.; Anh, L.H.; Minh, T.N. Phenolic compositions and antioxidant properties in bark, flower, inner skin, kernel and leaf extracts of Castanea crenata Sieb. et Zucc. Antioxidants 2017, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.L.; Hu, J.H.; Wang, L.; Kim, H.-S.; Jin, C.-W.; Cho, D.-H. Antioxidant and Anti-diabetes Activity of Extracts from Machilus thunbergii S. et Z. Korean J. Med. Crop Sci. 2010, 18, 34–39. [Google Scholar]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J. Chemom. 2014, 28, 623–632. [Google Scholar] [CrossRef]
- Azizan, A.; Lee, A.X.; Abdul Hamid, N.A.; Maulidiani, M.; Mediani, A.; Abdul Ghafar, S.Z.; Zolkeflee, N.K.Z.; Abas, F. Potentially bioactive metabolites from pineapple waste extracts and their antioxidant and α-glucosidase inhibitory activities by 1H NMR. Foods 2020, 9, 173. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.Y.; Seo, H.S.; Singh, D.; Lee, S.J.; Lee, C.H. Unraveling dynamic metabolomes underlying different maturation stages of berries harvested from Panax ginseng. J. Ginseng Res. 2020, 44, 413–423. [Google Scholar] [CrossRef]
- Lee, H.J.; Suh, D.H.; Jung, E.S.; Park, H.M.; Jung, G.-Y.; Do, S.-G.; Lee, C.H. Metabolomics of Lonicera caerulea fruit during ripening and its relationship with color and antioxidant activity. Food Res. Int. 2015, 78, 343–351. [Google Scholar] [CrossRef]
- Ren, J.; Zheng, Y.; Lin, Z.; Han, X.; Liao, W. Macroporous resin purification and characterization of flavonoids from Platycladus orientalis (L.) Franco and their effects on macrophage inflammatory response. Food Funct. 2017, 8, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.K.; Yang, H.; Yoon, J.S.; Jeong, E.J.; Ha, N.R.; Sung, S.H.; Kim, Y.C. Antifibrotic activity of diterpenes from Biota orientalis leaves on hepatic stellate cells. Arch. Pharmacal Res. 2008, 31, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Nazir, N.; Zahoor, M.; Nisar, M.; Khan, I.; Karim, N.; Abdel-Halim, H.; Ali, A. Phytochemical analysis and antidiabetic potential of Elaeagnus umbellata (Thunb.) in streptozotocin-induced diabetic rats: Pharmacological and computational approach. BMC Complementary Altern. Med. 2018, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Andrade, A.W.L.; da Conceição Machado, K.; da Conceição Machado, K.; Figueiredo, D.D.R.; David, J.M.; Islam, M.T.; Uddin, S.J.; Shilpi, J.A.; Costa, J.P. In vitro antioxidant properties of the biflavonoid agathisflavone. Chem. Cent. J. 2018, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Yoo, H.J.; Lee, M. Robustaflavone isolated from Nandina domestica using bioactivity-guided fractionation downregulates inflammatory mediators. Molecules 2019, 24, 1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Li, B.; Hou, Y.; Tian, Y.; Chen, L.; Liu, S.; Zhang, N.; Dong, J. Anti-inflammatory effects of chemical components from Ginkgo biloba L. male flowers on lipopolysaccharide-stimulated RAW264. 7 macrophages. Phytother. Res. 2019, 33, 989–997. [Google Scholar] [CrossRef]
- Kim, H.P.; Park, H.; Son, K.H.; Chang, H.W.; Kang, S.S. Biochemical pharmacology of biflavonoids: Implications for anti-inflammatory action. Arch. Pharmacal Res. 2008, 31, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Gontijo, V.S.; Dos Santos, M.H.; Viegas, C., Jr. Biological and chemical aspects of natural biflavonoids from plants: A brief review. Mini Rev. Med. Chem. 2017, 17, 834–862. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Hormaza, L.; Ramírez, A.M.; Quintero-Ortiz, C.; Cossio, M.; Medina, S.; Ferreres, F.; Gil-Izquierdo, A.; Osorio, E. Comprehensive characterization and antioxidant activities of the main biflavonoids of Garcinia madruno: A novel tropical species for developing functional products. J. Funct. Foods 2016, 27, 503–516. [Google Scholar] [CrossRef]
- Lee, M.Y.; Singh, D.; Kim, S.H.; Lee, S.J.; Lee, C.H. Ultrahigh pressure processing produces alterations in the metabolite profiles of Panax ginseng. Molecules 2016, 21, 816. [Google Scholar] [CrossRef] [Green Version]








| No. | Labeling | Family | Species | Parts | NIBR Number |
|---|---|---|---|---|---|
| 1 | Lar1 | Lardizabalaceae | Akebia quinata | stem | NIBRGR0000433376 |
| 2 | Lar2 | Lardizabalaceae | Akebia quinata | fruit | NIBRGR0000594502 |
| 3 | Ama1 | Amaryllidaceae | Allium tuberosum | etc. | NIBRGR0000612270 |
| 4 | Ama2 | Amaryllidaceae | Allium tuberosum | leaf, stem | NIBRGR0000612271 |
| 5 | Ama3 | Amaryllidaceae | Allium tuberosum | underground | NIBRGR0000612272 |
| 6 | Api1 | Apiaceae | Angelica dahurica | inflorescence | NIBRGR0000612861 |
| 7 | Ara1 | Araliaceae | Aralia cordata | stem | NIBRGR0000433525 |
| 8 | Ara2 | Araliaceae | Aralia cordata | leaf | NIBRGR0000594470 |
| 9 | Fag1 | Fagaceae | Castanea crenata | fruit | NIBRGR0000612236 |
| 10 | Ran1 | Ranunculaceae | Cimicifuga heracleifolia | aboveground | NIBRGR0000433787 |
| 11 | Ran2 | Ranunculaceae | Cimicifuga heracleifolia | underground | NIBRGR0000611752 |
| 12 | Com1 | Commelinaceae | Commelina communis | stem, leaf, flower, fruit | NIBRGR0000594517 |
| 13 | Ast1 | Asteraceae | Dendranthema boreale | stem, leaf | NIBRGR0000612853 |
| 14 | Ast2 | Asteraceae | Dendranthema indicum | stem, leaf, inflorescence | NIBRGR0000423023 |
| 15 | Ela1 | Elaeagnaceae | Elaeagnus umbellate | leaf | NIBRGR0000433417 |
| 16 | Ela2 | Elaeagnaceae | Elaeagnus umbellate | branch | NIBRGR0000595364 |
| 17 | Ara3 | Araliaceae | Eleutherococcus sessiliflorus | branch | NIBRGR0000433533 |
| 18 | Ara4 | Araliaceae | Eleutherococcus sessiliflorus | inflorescence | NIBRGR0000594494 |
| 19 | Ara5 | Araliaceae | Eleutherococcus sessiliflorus | leaf | NIBRGR0000597039 |
| 20 | Cel1 | Celastraceae | Euonymus alatus | branch | NIBRGR0000595199 |
| 21 | Lil1 | Liliaceae | Lilium lancifolium | root | NIBRGR0000597653 |
| 22 | Lil2 | Liliaceae | Lilium lancifolium | leaf, stem | NIBRGR0000597655 |
| 23 | Lil3 | Liliaceae | Lilium lancifolium | flower | NIBRGR0000610691 |
| 24 | Lil4 | Liliaceae | Lilium lancifolium | leaf, stem | NIBRGR0000616820 |
| 25 | Lau1 | Lauraceae | Lindera obtusiloba | branch | NIBRGR0000594799 |
| 26 | Lau2 | Lauraceae | Lindera obtusiloba | leaf | NIBRGR0000595318 |
| 27 | Lau3 | Lauraceae | Machilus thunbergia | branch | NIBRGR0000433461 |
| 28 | Lau4 | Lauraceae | Machilus thunbergia | leaf | NIBRGR0000595768 |
| 29 | Lam1 | Lamiaceae | Mentha arvensis | aboveground | NIBRGR0000612757 |
| 30 | Lam2 | Lamiaceae | Mentha arvensis | underground | NIBRGR0000612758 |
| 31 | Gra1 | Gramineae | Miscanthus sinensis | leaf, stem | NIBRGR0000595747 |
| 32 | Pin1 | Pinaceae | Pinus densiflora | branch | NIBRGR0000433437 |
| 33 | Pin2 | Pinaceae | Pinus densiflora | leaf | NIBRGR0000597148 |
| 34 | Pla1 | Plantaginaceae | Plantago asiatica | leaf, peduncle | NIBRGR0000611706 |
| 35 | Cup1 | Cupressaceae | Platycladus orientalis | fruit | NIBRGR0000433401 |
| 36 | Cup2 | Cupressaceae | Platycladus orientalis | branch | NIBRGR0000433615 |
| 37 | Cup3 | Cupressaceae | Platycladus orientalis | leaf | NIBRGR0000597151 |
| 38 | Sol1 | Solanaceae | Solanum nigrum | leaf, stem | NIBRGR0000433559 |
| 39 | Sol2 | Solanaceae | Solanum nigrum | leaf, stem, inflorescence | NIBRGR0000433569 |
| 40 | Fab1 | Fabaceae | Sophora flavescens | leaf | NIBRGR0000594794 |
| 41 | Fab2 | Fabaceae | Sophora flavescens | leaf, stem | NIBRGR0000594812 |
| 42 | Cuc1 | Cucurbitaceae | Trichosanthes kirilowii | leaf, stem | NIBRGR0000594804 |
| 43 | Cuc2 | Cucurbitaceae | Trichosanthes kirilowii | fruit | NIBRGR0000433378 |
| 44 | Cuc3 | Cucurbitaceae | Trichosanthes kirilowii | leaf | NIBRGR0000433567 |
| 45 | Cuc4 | Cucurbitaceae | Trichosanthes kirilowii | branch | NIBRGR0000433580 |
| 46 | Typ1 | Typhaceae | Typha orientalis | leaf, stem | NIBRGR0000595290 |
| 47 | Typ2 | Typhaceae | Typha orientalis | inflorescence | NIBRGR0000597721 |
| 48 | Rut1 | Rutaceae | Zanthoxylum schinifolium | leaf | NIBRGR0000408953 |
| 49 | Rut2 | Rutaceae | Zanthoxylum schinifolium | branch | NIBRGR0000594990 |
| 50 | Rut3 | Rutaceae | Zanthoxylum schinifolium | fruit | NIBRGR0000595216 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, S.R.; Lee, M.Y.; Kim, S.A.; Oh, J.; Hyun, D.W.; Lee, S.; Lee, B.-H.; Cho, J.Y.; Lee, C.H. Nontargeted Metabolomics as a Screening Tool for Estimating Bioactive Metabolites in the Extracts of 50 Indigenous Korean Plants. Metabolites 2021, 11, 585. https://doi.org/10.3390/metabo11090585
Choi SR, Lee MY, Kim SA, Oh J, Hyun DW, Lee S, Lee B-H, Cho JY, Lee CH. Nontargeted Metabolomics as a Screening Tool for Estimating Bioactive Metabolites in the Extracts of 50 Indigenous Korean Plants. Metabolites. 2021; 11(9):585. https://doi.org/10.3390/metabo11090585
Chicago/Turabian StyleChoi, Se Rin, Mee Youn Lee, Seung A Kim, Jieun Oh, Da Won Hyun, Sarah Lee, Byoung-Hee Lee, Jae Youl Cho, and Choong Hwan Lee. 2021. "Nontargeted Metabolomics as a Screening Tool for Estimating Bioactive Metabolites in the Extracts of 50 Indigenous Korean Plants" Metabolites 11, no. 9: 585. https://doi.org/10.3390/metabo11090585
APA StyleChoi, S. R., Lee, M. Y., Kim, S. A., Oh, J., Hyun, D. W., Lee, S., Lee, B.-H., Cho, J. Y., & Lee, C. H. (2021). Nontargeted Metabolomics as a Screening Tool for Estimating Bioactive Metabolites in the Extracts of 50 Indigenous Korean Plants. Metabolites, 11(9), 585. https://doi.org/10.3390/metabo11090585