Metabolomic Study of a Rat Model of Retinal Detachment
Abstract
:1. Introduction
2. Materials and Methods
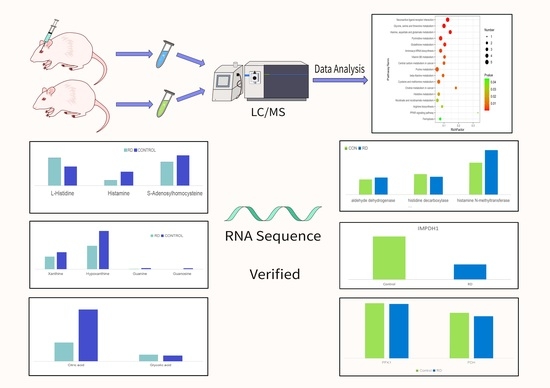
2.1. Animal Ethics and the Animal Model of Retinal Detachment
2.2. Sample Collection and Preparation
2.3. LC–MS/MS Analyses for Untargeted Metabolomics
2.4. Data Extraction and Analysis
2.5. RNA-Seq, Sequencing Data Extraction Analysis
2.6. Statistical Analysis
3. Results
3.1. Untargeted Metabolomics of Retina Samples of Rat Model of RD
3.2. Involved Pathways Related to Changed Metabolites in RD
3.2.1. Histidine Metabolism
3.2.2. Purine and Pyrimidine Metabolism
3.2.3. Phenylalanine, Tyrosine, and Tryptophan Biosynthesis
3.2.4. Glyoxylate and Dicarboxylate Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Steel, D. Retinal detachment. BMJ Clin. Evid. 2014, 2014, 0710. [Google Scholar]
- Kwok, J.M.; Yu, C.W.; Christakis, P.G. Retinal detachment. CMAJ 2020, 192, E312. [Google Scholar] [CrossRef] [Green Version]
- Schick, T.; Heimann, H.; Schaub, F. Retinal Detachment Part 1-Epidemiology, Risk Factors, Clinical Characteristics, Diagnostic Approach. Klin. Mon. Augenheilkd. 2020, 237, 1479–1491. [Google Scholar]
- Zhang, Z.-Y.; Sun, Y.-J.; Song, J.-Y.; Fan, B.; Li, G.-Y. Experimental models and examination methods of retinal detachment. Brain Res. Bull. 2021, 169, 51–62. [Google Scholar] [CrossRef]
- Yang, S.; Li, H.; Yao, H.; Zhang, Y.; Bao, H.; Wu, L.; Zhang, C.; Li, M.; Le Feng, L.; Zhang, J.; et al. Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy. Cell Death Differ. 2021, 28, 2351–2366. [Google Scholar] [CrossRef]
- Judge, A.; Dodd, M.S. Metabolism. Essays Biochem. 2020, 64, 607–647. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, H.; Jiang, P.; Liu, X.; Xu, D.; Wang, F. Investigating the pathological processes of rhegmatogenous retinal detachment and proliferative vitreoretinopathy with metabolomics analysis. Mol. BioSyst. 2014, 10, 1055–1062. [Google Scholar] [CrossRef]
- Pastor, J.C.; Rojas, J.; Pastor-Idoate, S.; Di Lauro, S.; Gonzalez-Buendia, L.; Delgado-Tirado, S. Proliferative vitreoretinopathy: A new concept of disease pathogenesis and practical consequences. Prog. Retin. Eye Res. 2016, 51, 125–155. [Google Scholar] [CrossRef]
- Yu, M.; Wu, Z.; Zhang, Z.; Huang, X.; Zhang, Q. Metabolomic Analysis of Human Vitreous in Rhegmatogenous Retinal Detachment Associated With Choroidal Detachment. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5706–5713. [Google Scholar] [CrossRef] [Green Version]
- Haines, N.R.; Manoharan, N.; Olson, J.L.; D’Alessandro, A.; Reisz, J.A. Metabolomics Analysis of Human Vitreous in Diabetic Retinopathy and Rhegmatogenous Retinal Detachment. J. Proteome Res. 2018, 17, 2421–2427. [Google Scholar] [CrossRef]
- Chidlow, G.; Chan, W.O.; Wood, J.P.M.; Casson, R.J. Differential Effects of Experimental Retinal Detachment on S- and M/L-Cones in Rats. Mol. Neurobiol. 2021, 59, 117–136. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Wilson, I.; Gika, H.G.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.P.; Holmes, E.; Nicholson, J. Global metabolic profiling procedures for urine using UPLC–MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Delyfer, M.-N.; Raffelsberger, W.; Mercier, D.; Korobelnik, J.-F.; Gaudric, A.; Charteris, D.G.; Tadayoni, R.; Metge, F.; Caputo, G.; Barale, P.-O.; et al. Transcriptomic Analysis of Human Retinal Detachment Reveals Both Inflammatory Response and Photoreceptor Death. PLoS ONE 2011, 6, e28791. [Google Scholar] [CrossRef]
- Yao, K.; Yu, Y.; Zhang, H. Construction for Long Non-Coding RNA (lncRNA)-Associated Competing Endogenous RNA (ceRNA) Network in Human Retinal Detachment (RD) with Proliferative Vitreoretinopathy (PVR). Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 26, e919871. [Google Scholar] [CrossRef]
- Panula, P.; Sundvik, M.; Karlstedt, K. Developmental roles of brain histamine. Trends Neurosci. 2014, 37, 159–168. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, R.; Hof, P.; O Duarte, R.; Moura, J.J.; Moura, I.; Liu, M.Y.; LeGall, J.; Hille, R.; Archer, M.; Romão, M.J. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc. Natl. Acad. Sci. USA 1996, 93, 8846–8851. [Google Scholar] [CrossRef] [Green Version]
- Crooks, J.; Kolb, H. Localization of GABA, glycine, glutamate and tyrosine hydroxylase in the human retina. J. Comp. Neurol. 1992, 315, 287–302. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, L.; Chen, H.; Chen, Y.Q.; Chen, W.; Song, Y.; Ratledge, C. Enhanced lipid accumulation in the yeast Yarrowia lipolytica by over-expression of ATP:citrate lyase from Mus musculus. J. Biotechnol. 2014, 192, 78–84. [Google Scholar] [CrossRef]
- Akimov, N.P.; Marshak, D.W.; Frishman, L.J.; Glickman, R.D.; Yusupov, R.G. Histamine reduces flash sensitivity of on ganglion cells in the primate retina. Investig. Ophthalmol. Vis. Sci. 2010, 51, 3825–3834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usui, T.; Kubo, Y.; Akanuma, S.; Hosoya, K. Β-alanine and l-histidine transport across the inner blood-retinal barrier: Potential involvement in L-carnosine supply. Exp. Eye Res. 2013, 113, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.C.S.; Martin, P.R.; Grünert, U. Topography of Neurons in the Rod Pathway of Human Retina. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2848–2859. [Google Scholar] [CrossRef] [Green Version]
- Hardie, R.C. A histamine-activated chloride channel involved in neurotransmission at a photoreceptor synapse. Nature 1989, 339, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; An, F.; Borycz, J.A.; Borycz, J.; Meinertzhagen, I.A.; Wang, T. Histamine Recycling Is Mediated by CarT, a Carcinine Transporter in Drosophila Photoreceptors. PLoS Genet. 2015, 11, e1005764. [Google Scholar] [CrossRef] [Green Version]
- Christiansen, J.R.; Pendse, N.D.; Kolandaivelu, S.; Bergo, M.O.; Young, S.G.; Ramamurthy, V. Deficiency of Isoprenylcysteine Carboxyl Methyltransferase (ICMT) Leads to Progressive Loss of Photoreceptor Function. J. Neurosci. 2016, 36, 5107–5114. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.A.; Granger, D.N. Xanthine oxidase: Biochemistry, distribution and physiology. Acta Physiol. Scand. Suppl. 1986, 548, 87–99. [Google Scholar]
- Fox, N.E.; van Kuijk, F.J. Immunohistochemical localization of xanthine oxidase in human retina. Free Radic. Biol. Med. 1998, 24, 900–905. [Google Scholar] [CrossRef]
- Tasca, C.I.; Lanznaster, D.; Oliveira, K.A.; Fernández-Dueñas, V.; Ciruela, F. Neuromodulatory Effects of Guanine-Based Purines in Health and Disease. Front. Cell. Neurosci. 2018, 12, 376. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; French, J.B.; Fang, Y.; Benkovic, S.J. The purinosome, a multi-protein complex involved in the de novo biosynthesis of purines in humans. Chem. Commun. 2013, 49, 4444–4452. [Google Scholar] [CrossRef] [Green Version]
- Plana-Bonamaisó, A.; López-Begines, S.; Fernández-Justel, D.; Junza, A.; Soler-Tapia, A.; Andilla, J.; Loza-Alvarez, P.; Rosa, J.L.; Miralles, E.; Casals, I.; et al. Post-translational regulation of retinal IMPDH1 in vivo to adjust GTP synthesis to illumination conditions. Elife 2020, 9, e56418. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Bendjelloul, F.; Ballerini, P.; D’Alimonte, I.; Nargi, E.; Jiang, C.; Huang, X.; Rathbone, M.P. Guanosine reduces apoptosis and inflammation associated with restoration of function in rats with acute spinal cord injury. Purinergic Signal. 2007, 3, 411–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, A.H. Identification of tyrosine kinase Trk as a nerve growth factor receptor. Cell Regul. 1991, 2, 685–690. [Google Scholar] [CrossRef]
- Iwashita, S.; Kobayashi, M. Signal transduction system for growth factor receptors associated with tyrosine kinase activity: Epidermal growth factor receptor signalling and its regulation. Cell. Signal. 1992, 4, 123–132. [Google Scholar] [CrossRef]
- Rios, M.; Habecker, B.; Sasaoka, T.; Eisenhofer, G.; Tian, H.; Landis, S.; Chikaraishi, D.; Roffler-Tarlov, S. Catecholamine Synthesis is Mediated by Tyrosinase in the Absence of Tyrosine Hydroxylase. J. Neurosci. 1999, 19, 3519–3526. [Google Scholar] [CrossRef] [Green Version]
- Hu, D.-N.; Simon, J.D.; Sarna, T. Role of Ocular Melanin in Ophthalmic Physiology and Pathology. Photochem. Photobiol. 2008, 84, 639–644. [Google Scholar] [CrossRef]
- Cherix, A.; Brodier, L.; Poitry-Yamate, C.; Matter, J.-M.; Gruetter, R. The Appearance of the Warburg Effect in the Developing Avian Eye Characterized In Ovo: How Neurogenesis Can Remodel Neuroenergetics. Investig. Opthalmology Vis. Sci. 2020, 61, 3. [Google Scholar] [CrossRef]
- Ganetzky, R.; McCormick, E.M.; Falk, M.J. Primary Pyruvate Dehydrogenase Complex Deficiency Overview; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Sun, J.; Aluvila, S.; Kotaria, R.; Mayor, J.A.; Walters, D.E.; Kaplan, R.S. Mitochondrial and Plasma Membrane Citrate Transporters: Discovery of Selective Inhibitors and Application to Structure/Function Analysis. Mol. Cell. Pharmacol. 2010, 2, 101–110. [Google Scholar]
- Curcio, C.A.; Allen, K.A.; Sloan, K.R.; Lerea, C.L.; Hurley, J.B.; Klock, I.B.; Milam, A.H. Distribution and morphology of human cone photoreceptors stained with anti-blue opsin. J. Comp. Neurol. 1991, 312, 610–624. [Google Scholar] [CrossRef]
- Mollon, J.D.; Regan, B.C.; Bowmaker, J.K. What is the function of the cone-rich rim of the retina? Eye 1998, 12 Pt 3b, 548–552. [Google Scholar] [CrossRef]







| Metabolite | VIP | a Fold Change | p-Value | mz | rt |
|---|---|---|---|---|---|
| Sarcosine | 1.588794734 | 0.753842042 | 0.00172 | 90.05489512 | 43.3375 |
| Cytidine | 1.809324897 | 0.460435551 | 0.00003 | 244.0867888 | 56.1297 |
| 2-Pyrrolidinone | 2.031331915 | 0.671816243 | 0.00004 | 86.06002251 | 44.74485 |
| Nb-p-Coumaroyltryptamine | 1.905917252 | 7.582405045 | 0.02603 | 307.1416345 | 496.676 |
| Guanosine | 2.153682946 | 0.198874723 | 0.00062 | 284.0989995 | 154.511 |
| Uridine | 1.64987382 | 0.580089132 | 0.00488 | 245.0759403 | 92.4567 |
| L-Carnitine | 1.698905167 | 1.751054619 | 0.00012 | 162.1079781 | 42.6997 |
| Coriandrone D | 1.453741941 | 1.460356861 | 0.03866 | 353.1589536 | 910.33 |
| Uracil | 1.690394112 | 0.613961856 | 0.00439 | 113.0335019 | 92.8533 |
| 2-Hydroxypyridine | 1.812738744 | 0.863836807 | 0.00079 | 96.04164293 | 69.8369 |
| Ascorbic acid | 2.04657221 | 0.57252687 | 0.00005 | 177.0330868 | 51.13885 |
| 3-Hydroxy-2-methylpyridine-4,5-dicarboxylate | 1.814382779 | 0.475699949 | 0.00003 | 198.0372243 | 64.1571 |
| L-Histidine | 1.529516838 | 1.44624588 | 0.00052 | 156.0754384 | 39.2151 |
| Niacinamide | 1.341138793 | 0.735454619 | 0.00428 | 123.0552253 | 70.6486 |
| Propionylcarnitine | 1.366650349 | 2.162667902 | 0.00362 | 218.1380477 | 136.899 |
| Pyroglutamic acid | 1.825841104 | 0.403212341 | 0.00005 | 130.0498196 | 82.9935 |
| Cytosine | 1.783710152 | 0.47980553 | 0.00003 | 112.050413 | 57.1205 |
| 4-Methoxybrassinin | 1.659923965 | 0.516514641 | 0.00334 | 267.0587392 | 91.8458 |
| Guanine | 2.162064502 | 0.208577348 | 0.00081 | 152.0564281 | 154.536 |
| Hypoxanthine | 1.725128074 | 0.611006413 | 0.00005 | 137.0466486 | 70.23255 |
| m-Xylene | 1.921696126 | 1.243960894 | 0.00133 | 107.0826798 | 939.389 |
| Xanthine | 1.457026673 | 0.743827737 | 0.00145 | 153.0433249 | 79.82625 |
| Phosphorylcholine | 1.311159946 | 1.419536743 | 0.00481 | 184.0697118 | 40.3446 |
| L-Palmitoylcarnitine | 1.680174185 | 4.978528439 | 0.03278 | 400.3412165 | 964.8555 |
| Succinyladenosine | 1.480016326 | 0.618517209 | 0.00201 | 384.1150194 | 330.112 |
| (-)-Epigallocatechin | 1.281914517 | 2.687838842 | 0.01162 | 307.0833389 | 105.983 |
| gamma-Glutamylcysteine | 1.16082442 | 0.633360213 | 0.02104 | 251.0704476 | 68.27515 |
| N2-gamma-Glutamylglutamine | 1.79789316 | 0.502733902 | 0.00008 | 276.1224994 | 44.7747 |
| 2-(Methylthio)propane | 1.907264524 | 1.267761325 | 0.00037 | 91.05418475 | 939.711 |
| Dimethyl dialkyl ammonium chloride | 1.363211764 | 1.178980467 | 0.04779 | 304.3001743 | 940.381 |
| S-Adenosylhomocysteine | 1.152857028 | 0.674020148 | 0.00332 | 385.1285023 | 110.14 |
| LysoPC(22:6(4Z, 7Z, 10Z, 13Z, 16Z, 19Z)) | 1.289484261 | 0.498294474 | 0.01125 | 568.3387229 | 984.4805 |
| Atroviridin | 1.909820405 | 0.393778382 | 0.00000 | 327.0798169 | 100.4 |
| Oxidized glutathione | 1.188118624 | 4.000915552 | 0.02011 | 613.1586921 | 105.3015 |
| Guggulsterone | 1.38032097 | 1.641203242 | 0.04156 | 343.2244804 | 972.79 |
| (1R, 2R, 4R, 8S)-p-Menthane-1,2,8,9-tetrol 9-glucoside | 1.248626769 | 0.575134248 | 0.00270 | 367.1949551 | 1013.06 |
| Ambonic acid | 1.624811991 | 2.388539493 | 0.00620 | 469.3625627 | 1112.09 |
| Butyrylcarnitine | 1.58194961 | 2.910638976 | 0.00087 | 232.154252 | 279.752 |
| cis-4-Carboxymethylenebut-2-en-4-olide | 1.746288625 | 0.692541355 | 0.01425 | 141.0175682 | 51.75935 |
| Methyl phenyl disulfide | 1.260869035 | 0.681308476 | 0.00151 | 157.0143964 | 51.5829 |
| 2-Methylbutyroylcarnitine | 1.389115898 | 2.092890713 | 0.00103 | 246.1699963 | 402.919 |
| Creatine | 1.75441836 | 0.671404514 | 0.00078 | 132.076239 | 43.8692 |
| N-Acetylornithine | 1.538467126 | 1.376210914 | 0.00205 | 175.1132487 | 38.8724 |
| Vitamin A | 1.18216123 | 0.434496736 | 0.00634 | 269.2288829 | 1010.94 |
| 1-deoxy-1-(N6-lysino)-D-fructose | 1.772390317 | 0.665660902 | 0.00050 | 134.0442436 | 45.63555 |
| 2-(2-Furanyl)-3-piperidinol | 2.015285296 | 10.37822452 | 0.03335 | 168.1016873 | 97.7082 |
| LysoPE(22:6(4Z, 7Z, 10Z, 13Z, 16Z, 19Z)/0:0) | 1.68145778 | 0.420387817 | 0.00106 | 526.2975318 | 985.298 |
| gamma-Glutamylglutamic acid | 2.17322362 | 0.344750109 | 0.00000 | 277.1105039 | 53.96735 |
| 5’-Deoxy-5’-(methylsulfinyl)adenosine | 1.432320977 | 0.49083823 | 0.00000 | 314.0810526 | 46.68055 |
| Brassicanal C | 1.639967821 | 0.402276508 | 0.00003 | 224.0375107 | 70.74 |
| alpha-Methylstyrene | 1.049964179 | 1.497412492 | 0.02551 | 119.0854447 | 939.6095 |
| 3-O-Acetylepisamarcandin | 1.31455005 | 1.62786731 | 0.01162 | 460.2694428 | 939.137 |
| Elaidic carnitine | 1.522562393 | 3.678436325 | 0.00832 | 426.3562901 | 966.6665 |
| L-Aspartyl-4-phosphate | 2.045673216 | 0.473543833 | 0.00000 | 214.0126044 | 54.0449 |
| Persicaxanthin | 1.166835004 | 0.641843353 | 0.02979 | 385.2810119 | 985.3045 |
| Prostaglandin G2 | 1.361188168 | 0.605822078 | 0.00202 | 351.2220203 | 1012.245 |
| Histamine | 1.11300757 | 0.385722377 | 0.00023 | 96.92917902 | 39.3631 |
| Lansoprazole sulfone | 1.575610946 | 0.447892561 | 0.01350 | 386.0851118 | 92.13835 |
| Cysteic acid | 1.67467486 | 0.658755513 | 0.00010 | 170.0084242 | 39.9788 |
| MS2 Name | VIP | a Fold Change | p-Value | mz | rt |
|---|---|---|---|---|---|
| Glycine | 1.308701658 | 1.418244394 | 0.00497 | 74.02476574 | 41.604 |
| Pyrrolidonecarboxylic acid | 1.182346811 | 0.73594283 | 0.02202 | 128.0353476 | 45.9248 |
| L-Norleucine | 1.299366092 | 1.359856849 | 0.00721 | 130.0870664 | 85.1877 |
| Dodecanoic acid | 1.013687543 | 1.112895178 | 0.02735 | 199.1699751 | 998.35 |
| Pentadecanoic acid | 1.572438882 | 1.238311403 | 0.00004 | 241.2170986 | 1032.37 |
| Capric acid | 1.22167135 | 1.160060897 | 0.00631 | 171.1355328 | 957.741 |
| Linoleic acid | 1.093421764 | 1.201474709 | 0.04900 | 279.2325558 | 1036.41 |
| O-Phosphoethanolamine | 1.016072298 | 1.216167453 | 0.01543 | 140.0101022 | 37.523 |
| Adenine | 1.446337373 | 1.629672015 | 0.01034 | 134.0472006 | 133.03 |
| L-Tyrosine | 1.272211932 | 1.527389323 | 0.00934 | 180.0663514 | 71.8062 |
| Pyridoxal | 1.319697042 | 1.421159579 | 0.00678 | 166.0464366 | 108.894 |
| Sarcosine | 1.422117232 | 0.761449281 | 0.00772 | 88.04041793 | 41.2361 |
| L-Phenylalanine | 1.429734239 | 1.42959732 | 0.00585 | 164.0714753 | 191.467 |
| N-Acetyl-L-aspartic acid | 1.738817993 | 0.5753958 | 0.00001 | 174.0405148 | 36.9303 |
| 13S-hydroxyoctadecadienoic acid | 1.198241881 | 1.17631196 | 0.01217 | 295.2268903 | 984.563 |
| D-Glutamine | 1.213915884 | 0.729900743 | 0.00510 | 145.0688397 | 39.775 |
| L-Aspartic acid | 1.280263363 | 1.387046842 | 0.00218 | 132.0300608 | 37.6211 |
| Glycolic acid | 1.247166563 | 1.151900036 | 0.00991 | 75.00868616 | 1165.97 |
| Xanthosine | 1.754820236 | 2.161916582 | 0.00002 | 283.0677416 | 231.952 |
| 5,6-DHET | 1.076984772 | 1.17425398 | 0.01023 | 337.2377561 | 1000.645 |
| 16-Hydroxy hexadecanoic acid | 1.047890336 | 0.864921063 | 0.01094 | 271.2201796 | 982.637 |
| Myristoleic acid | 1.390731089 | 1.52634857 | 0.00146 | 225.1854497 | 1008.66 |
| 16(R)-HETE | 1.010988944 | 2.271573423 | 0.04334 | 319.2267346 | 990.646 |
| Ascorbic acid | 1.18946295 | 0.749711824 | 0.03688 | 175.0256936 | 38.9044 |
| Deoxyinosine | 1.91372124 | 3.310838197 | 0.00041 | 251.0803053 | 193.8595 |
| 5Z-Dodecenoic acid | 1.85788377 | 1.465643497 | 0.00000 | 197.1541993 | 981.979 |
| Leukotriene B4 | 1.141316937 | 1.57686519 | 0.04224 | 335.2199433 | 974.804 |
| Citric acid | 1.334618831 | 0.362946196 | 0.00560 | 191.0191466 | 36.623 |
| D-Myo-inositol 4-phosphate | 1.329663611 | 2.419328351 | 0.02151 | 259.0210802 | 36.9303 |
| S-Adenosylhomocysteine | 1.349938422 | 0.779341895 | 0.00077 | 383.1136101 | 236.586 |
| Thymidine | 1.700378459 | 2.107103442 | 0.00024 | 241.0778921 | 256.103 |
| Sorbitol | 1.250229055 | 1.489605484 | 0.01152 | 181.0673795 | 71.9667 |
| (10E, 12Z)-(9S)-9-Hydroperoxyoctadeca-10,12-dienoic acid | 1.020361506 | 1.198479736 | 0.02153 | 311.2223869 | 966.673 |
| Ethylparaben | 1.733042441 | 23.16190628 | 0.01128 | 165.0511808 | 789.595 |
| Guanosine | 1.809686128 | 0.283277591 | 0.00120 | 282.0842135 | 171.802 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
She, X.; Zhou, Y.; Liang, Z.; Wei, J.; Xie, B.; Zhang, Y.; Shen, L. Metabolomic Study of a Rat Model of Retinal Detachment. Metabolites 2022, 12, 1077. https://doi.org/10.3390/metabo12111077
She X, Zhou Y, Liang Z, Wei J, Xie B, Zhang Y, Shen L. Metabolomic Study of a Rat Model of Retinal Detachment. Metabolites. 2022; 12(11):1077. https://doi.org/10.3390/metabo12111077
Chicago/Turabian StyleShe, Xiangjun, Yifan Zhou, Zhi Liang, Jin Wei, Bintao Xie, Yun Zhang, and Lijun Shen. 2022. "Metabolomic Study of a Rat Model of Retinal Detachment" Metabolites 12, no. 11: 1077. https://doi.org/10.3390/metabo12111077
APA StyleShe, X., Zhou, Y., Liang, Z., Wei, J., Xie, B., Zhang, Y., & Shen, L. (2022). Metabolomic Study of a Rat Model of Retinal Detachment. Metabolites, 12(11), 1077. https://doi.org/10.3390/metabo12111077