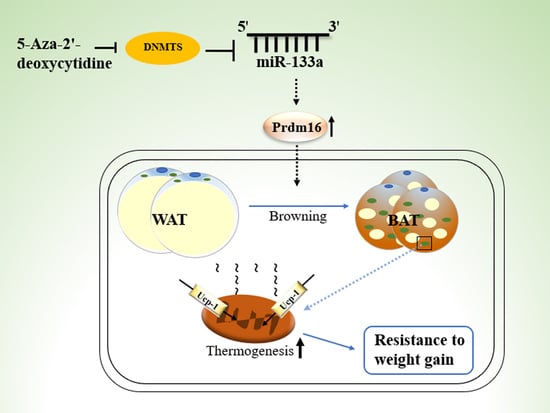
5-Aza-2′-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Determination of Parameters
2.3. Cells
2.4. Oil Red O Staining
2.5. Determination of 5-Methyl Cytosine in Adipose Tissue
2.6. Determination of Serum Leptin Levels
2.7. Histology and Immunofluorescence
2.8. Western Blotting
2.9. RT-PCR Analysis
2.10. MicroRNA Transfection of Primary Adipocytes
2.11. Statistical Analysis
3. Results
3.1. DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Influenced Normal Growth Indicators in Mice
3.2. Adipose Tissue of Mice Exhibits Browning Characteristics after 5-Aza-dC Treatment
3.3. 5-Aza-dC Promotes the Browning of WAT by Regulating Related Factors In Vivo
3.4. 5-Aza-dC Inhibited the Accumulation of Lipid Droplets and Promoted the Browning of Adipocytes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rosen, E.D.; Spiegelman, B.M. What we talk about when we talk about fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lasar, D.; Julius, A.; Fromme, T.; Klingenspor, M. Browning attenuates murine white adipose tissue expansion during postnatal development. Biochim. Biophys. Acta 2013, 1831, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Lidell, M.E.; Betz, M.J.; Enerback, S. Two types of brown adipose tissue in humans. Adipocyte 2014, 3, 63–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaisanlahti, A.; Glumoff, T. Browning of white fat: Agents and implications for beige adipose tissue to type 2 diabetes. J. Physiol. Biochem. 2019, 75, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP-1containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar]
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat. Rev. Mol. Cell Biol. 2016, 17, 480–495. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Page, A.J.; Hutchison, A.T.; Wittert, G.; Heilbronn, L.K. Intermittent fasting increases energy expenditure and promotes adipose tissue browning in mice. Nutrition 2019, 66, 38–43. [Google Scholar] [CrossRef]
- Montanari, T.; Pošćić, N.; Colitti, M. Factors involved in white-to-brown adipose tissue conversion and in thermogenesis: A review. Obes. Rev. 2017, 18, 495–513. [Google Scholar] [CrossRef]
- Waalen, J. The genetics of human obesity. Transl. Res. 2014, 164, 293–301. [Google Scholar] [CrossRef]
- Andrews, S.V.; Sheppard, B.; Windham, G.C.; Schieve, L.A.; Schendel, D.E.; Croen, L.A.; Chopra, P.; Alisch, R.S.; Newschaffer, C.J.; Warren, S.T.; et al. Case-control meta-analysis of blood DNA methylation and autism spectrum disorder. Mol. Autism 2018, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Lim, Y.C.; Chia, S.Y.; Jin, S.; Han, W.; Ding, C.; Sun, L. Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol. Metab. 2016, 5, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Wu, R.; Yang, X.; Kou, S.; MacDougald, O.A.; Yu, L.; Shi, H.; Xue, B. Inhibiting DNA methylation switches adipogenesis to osteoblast to genesis by activating Wnt10a. Sci. Rep. 2016, 6, 25283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, H.; Kogo, Y.; Ohgane, J.; Hattori, N.; Yagi, S.; Tanaka, S.; Shiota, K. Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2008, 366, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Shore, A.; Karamitri, A.; Kemp, P.; Speakman, J.R.; Lomax, M.A. Role of Ucp1 enhancer methylation and chromatin remodelling in the control of Ucp1 expression in murine adipose tissue. Diabetologia 2010, 53, 1164–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Raghavan, P.; Thomou, T.; Boucher, J.; Robida-Stubbs, S.; Macotela, Y.; Russell, S.J.; Kirkland, J.L.; Blackwell, T.K.; Kahn, C.R. Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metab. 2012, 16, 336–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, W.; Nie, Y.; Qaher, M.; Horton, H.E.; Yue, F.; Asakura, A.; Kuang, S. Loss of MyoD Promotes Fate Trans differentiation of Myoblasts Into Brown Adipocytes. eBioMedicine 2017, 16, 212–223. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Pasut, A.; Soleimani, V.D.; Bentzinger, C.F.; Antoun, G.; Thorn, S.; Seale, P.; Fernando, P.; van Ijcken, W.; Grosveld, F.; et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013, 17, 210–224. [Google Scholar] [CrossRef] [Green Version]
- Gao, R.; Chen, W.; Yan, H.; Xie, X.; Liu, D.; Wu, C.; Zhu, Z.; Li, H.; Dong, F.; Wang, L. PPARγ agonist rosiglitazone switches fuel preference to lipids in promoting thermogenesis under cold exposure in C57BL/6 mice. J. Proteom. 2018, 176, 24–36. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef] [PubMed]
- Arndt, A.-K.; Schafer, S.; Drenckhahn, J.-D.; Sabeh, M.K.; Plovie, E.R.; Caliebe, A.; Klopocki, E.; Musso, G.; Werdich, A.A.; Kalwa, H.; et al. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am. J. Hum. Genet. 2013, 93, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Kuryłowicz, A.; Puzianowska-Kuźnicka, M. Induction of Adipose Tissue Browning as a Strategy to Combat Obesity. Int. J. Mol. Sci. 2020, 21, 6241. [Google Scholar] [CrossRef] [PubMed]
- Gulyaeva, O.; Dempersmier, J.; Sul, H.S. Genetic and epigenetic control of adipose development. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 3–12. [Google Scholar] [CrossRef]
- Kahn, B.B.; Flier, J.S. Obesity and insulin resistance. J. Clin. Investig. 2000, 106, 473–481. [Google Scholar] [CrossRef] [Green Version]
- Boi, S.K.; Buchta, C.M.; Pearson, N.A.; Francis, M.B.; Meyerholz, D.; Grobe, J.; Norian, L.A. Obesity alters immune and metabolic profiles: New insight from obese-resistant mice on high-fat diet. Obesity 2016, 24, 2140–2149. [Google Scholar] [CrossRef]
- Bunyan, J.; Murrell, E.A.; Shah, P.P. The induction of obesity in rodents by means of monosodium glutamate. Br. J. Nutr. 1976, 35, 25–39. [Google Scholar] [CrossRef]
- Giralt, M.; Villarroya, F. White, brown, beige/brite: Different adipose cells for different functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef] [Green Version]
- D’Alessio, A.C.; Weaver, I.C.; Szyf, M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol. Cell Biol. 2007, 27, 7462–7474. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scimè, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fan, X.; Yan, J.; Chen, M.; Zhu, M.; Tang, Y.; Liu, S.; Tang, Z. A comprehensive epigenome atlas reveals DNA methylation regulating skeletal muscle development. Nucleic Acids Res. 2021, 49, 1313–1329. [Google Scholar] [CrossRef] [PubMed]
- Seaborne, R.A.; Strauss, J.; Cocks, M.; Shepherd, S.; O’Brien, T.D.; Van Someren, K.A.; Bell, P.G.; Murgatroyd, C.; Morton, J.P.; Stewart, C.E.; et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018, 8, 1898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voisin, S.; Harvey, N.R.; Haupt, L.M.; Griffiths, L.; Ashton, K.; Coffey, V.G.; Doering, T.; Thompson, J.-L.; Benedict, C.; Cedernaes, J.; et al. An epigenetic clock for human skeletal muscle. J. Cachexia Sarcopenia Muscle 2020, 11, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef]
- Howlett, K.F.; McGee, S.L. Epigenetic regulation of skeletal muscle metabolism. Clin. Sci. 2016, 130, 1051–1063. [Google Scholar] [CrossRef]
- Sun, L.; Xie, H.; Mori, M.A.; Alexander, R.; Yuan, B.; Hattangadi, S.M.; Liu, Q.; Kahn, C.R.; Lodish, H.F. Mir193b-365 is essential for brown fat differentiation. Nat. Cell Biol. 2011, 13, 958–965. [Google Scholar] [CrossRef] [Green Version]
- Mori, M.; Nakagami, H.; Rodriguez-Araujo, G.; Nimura, K.; Kaneda, Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012, 10, e1001314. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Bi, P.; Shan, T.; Yang, X.; Yin, H.; Wang, Y.-X.; Liu, N.; Rudnicki, M.A.; Kuang, S. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013, 9, e1003626. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, J.; Jia, Y.; Yu, H.; Yan, H.; Shen, Q.; Xu, Y.; Li, Y.; Yang, M. 5-Aza-2′-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16. Metabolites 2022, 12, 1131. https://doi.org/10.3390/metabo12111131
Liang J, Jia Y, Yu H, Yan H, Shen Q, Xu Y, Li Y, Yang M. 5-Aza-2′-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16. Metabolites. 2022; 12(11):1131. https://doi.org/10.3390/metabo12111131
Chicago/Turabian StyleLiang, Jia, Ying Jia, Huixin Yu, Haijing Yan, Qingyu Shen, Yong Xu, Yana Li, and Meizi Yang. 2022. "5-Aza-2′-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16" Metabolites 12, no. 11: 1131. https://doi.org/10.3390/metabo12111131
APA StyleLiang, J., Jia, Y., Yu, H., Yan, H., Shen, Q., Xu, Y., Li, Y., & Yang, M. (2022). 5-Aza-2′-Deoxycytidine Regulates White Adipocyte Browning by Modulating miRNA-133a/Prdm16. Metabolites, 12(11), 1131. https://doi.org/10.3390/metabo12111131