Pudding Proteomics: Cyclomaltodextrin Glucanotransferase and Microbial Proteases Can Liquefy Extended Shelf Life Dairy Products
Abstract
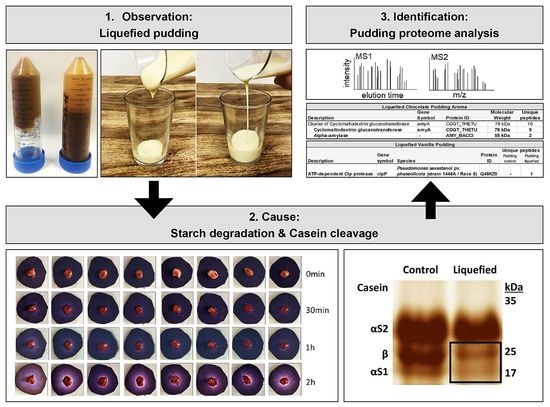
:1. Introduction
2. Results
2.1. Identification and Further Characterization of Added Amylases in Yoghurt
2.2. β-Casein Cleavage by Amylase Preparation from B. amyloliquefaciens
2.3. Identification of Cyclomaltodextrin Glucanotransferase from the Aroma of Liquefied Pudding
2.4. Detection of Protease from Pseudomonas Contamination in Liquefied Pudding
3. Discussion
4. Materials and Methods
4.1. Dairy Products, Casein, and Amylase Preparations
4.2. 1D and 2D SDS-PAGE
4.3. Zymography
4.4. Sample Digestion for Differential Proteome Analysis
4.5. Mass Spectrometric Analysis and Protein Identification
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rysstad, G.; Kolstad, J. Extended shelf life milk-advances in technology. Int. J. Dairy Technol. 2006, 59, 85–96. [Google Scholar] [CrossRef]
- Schmidt, V.S.; Kaufmann, V.; Kulozik, U.; Scherer, S.; Wenning, M. Microbial biodiversity, quality and shelf life of microfiltered and pasteurized extended shelf life (ESL) milk from Germany, Austria and Switzerland. Int. J. Food Microbiol. 2012, 154, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Paul, J.S.; Gupta, N.; Beliya, E.; Tiwari, S.; Jadhav, S.K. Aspects and Recent Trends in Microbial alpha-Amylase: A Review. Appl Biochem. Biotechnol. 2021, 193, 2649–2698. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Satyanarayana, T. Bacterial and Archaeal alpha-Amylases: Diversity and Amelioration of the Desirable Characteristics for Industrial Applications. Front. Microbiol. 2016, 7, 1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, A.; Nigam, P.; Soccol, C.R.; Soccol, V.T.; Singh, D.; Mohan, R. Advances in microbial amylases. Biotechnol. Appl. Biochem. 2000, 31, 135–152. [Google Scholar] [CrossRef] [PubMed]
- de Souza, P.M.; de Oliveira Magalhaes, P. Application of microbial alpha-amylase in industry—A review. Braz. J. Microbiol. 2010, 41, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Ezernieks, V.; Elkins, A.; Nguyen, N.; Moate, P.J.; Cocks, B.G.; Rochfort, S. Milk Bottom-Up Proteomics: Method Optimization. Front. Genet. 2015, 6, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geyer, P.E.; Holdt, L.M.; Teupser, D.; Mann, M. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 2017, 13, 942. [Google Scholar] [CrossRef] [PubMed]
- Roncada, P.; Piras, C.; Soggiu, A.; Turk, R.; Urbani, A.; Bonizzi, L. Farm animal milk proteomics. J. Proteomics 2012, 75, 4259–4274. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Geurts, N.; Martens, E.; Van den Steen, P.E.; Opdenakker, G. Zymography methods for visualizing hydrolytic enzymes. Nat. Methods 2013, 10, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Stam, M.R.; Danchin, E.G.; Rancurel, C.; Coutinho, P.M.; Henrissat, B. Dividing the large glycoside hydrolase family 13 into subfamilies: Towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 2006, 19, 555–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centeno-Leija, S.; Espinosa-Barrera, L.; Velazquez-Cruz, B.; Cardenas-Conejo, Y.; Virgen-Ortiz, R.; Valencia-Cruz, G.; Saenz, R.A.; Marin-Tovar, Y.; Gomez-Manzo, S.; Hernandez-Ochoa, B.; et al. Mining for novel cyclomaltodextrin glucanotransferases unravels the carbohydrate metabolism pathway via cyclodextrins in Thermoanaerobacterales. Sci. Rep. 2022, 12, 730. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.H.; Kim, J.S.; Jeong, H.M.; Shin, Y.J.; Hong, J.S.; Choi, H.D.; Shim, J.H. Development of Freeze-Thaw Stable Starch through Enzymatic Modification. Foods 2021, 10, 2269. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.F.; Zhang, J.Y.; Sun, Q.; Wang, M.; Gu, Z.B.; Du, G.C.; Wu, J.; Chen, J. Mutations of Lysine 47 in cyclodextrin glycosyltransferase from Paenibacillus macerans enhance beta-cyclodextrin specificity. J. Agric. Food. Chem. 2009, 57, 8386–8391. [Google Scholar] [CrossRef] [PubMed]
- Jurczak, A.; Jamka-Kasprzyk, M.; Bębenek, Z.; Staszczyk, M.; Jagielski, P.; Kościelniak, D.; Gregorczyk-Maga, I.; Kołodziej, I.; Kępisty, M.; Kukurba-Setkowicz, M.; et al. Differences in Sweet Taste Perception and Its Association with the Streptococcus mutans Cariogenic Profile in Preschool Children with Caries. Nutrients 2020, 12, 2592. [Google Scholar] [CrossRef]
- Mihaylova, D.; Popova, A.; Goranova, Z.; Petkova, D.; Doykina, P.; Lante, A. The Perspective of Nectarine Fruit as a Sugar Substituent in Puddings Prepared with Corn and Rice Starch. Foods 2021, 10, 2563. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Enzymes, pseudoenzymes, and moonlighting proteins: Diversity of function in protein superfamilies. FEBS J. 2020, 287, 4141–4149. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins. Trends. Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins: Old proteins learning new tricks. Trends. Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins—Nature’s Swiss army knives. Sci. Prog. 2017, 100, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zabad, S.; Liu, H.; Wang, W.; Jeffery, C. MoonProt 2.0: An expansion and update of the moonlighting proteins database. Nucleic Acids Res. 2018, 46, D640–D644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjani, V.; Janeček, S.; Chai, K.P.; Shahir, S.; Abdul Rahman, R.N.; Chan, K.G.; Goh, K.M. Protein engineering of selected residues from conserved sequence regions of a novel Anoxybacillus α-amylase. Sci. Rep. 2014, 4, 5850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Kaaij, R.M.; Janeček, Š.; van der Maarel, M.; Dijkhuizen, L. Phylogenetic and biochemical characterization of a novel cluster of intracellular fungal alpha-amylase enzymes. Microbiology 2007, 153, 4003–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janeček, Š.; Svensson, B.; MacGregor, E.A. α-Amylase: An enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci. 2014, 71, 1149–1170. [Google Scholar] [CrossRef]
- Wang, Y.; Li, F.; Zhang, Y. Preliminary investigation on the action modes of an oligosaccharide-producing multifunctional amylase. Appl. Biochem. Biotechnol. 2010, 160, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Satyanarayana, T. Dimerization mediates thermo-adaptation, substrate affinity and transglycosylation in a highly thermostable maltogenic amylase of Geobacillus thermoleovorans. PLoS ONE 2013, 8, e73612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Zhu, X.; Li, Y.; Cao, H.; Zhang, Y. Functional characterization of a special thermophilic multifunctional amylase OPMA-N and its N-terminal domain. Acta. Biochim. Biophys. Sin. 2011, 43, 324–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsunasawa, S.; Masaki, T.; Hirose, M.; Soejima, M.; Sakiyama, F. The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. J. Biol. Chem. 1989, 264, 3832–3839. [Google Scholar] [CrossRef]
- Slifkin, M.; Cumbie, R. Serogrouping single colonies of beta-hemolytic streptococci with achromopeptidase extraction. J. Clin. Microbiol. 1987, 25, 1555–1556. [Google Scholar] [CrossRef] [Green Version]
- Biwer, A.; Antranikian, G.; Heinzle, E. Enzymatic production of cyclodextrins. Appl. Microbiol. Biotechnol. 2002, 59, 609–617. [Google Scholar] [CrossRef]
- Gonzalez Pereira, A.; Carpena, M.; Garcia Oliveira, P.; Mejuto, J.C.; Prieto, M.A.; Simal Gandara, J. Main Applications of Cyclodextrins in the Food Industry as the Compounds of Choice to Form Host-Guest Complexes. Int. J. Mol. Sci. 2021, 22, 2339. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.H.; Rasti, B.; Sulistyo, J.; Hamid, M.A. Comprehensive study on transglycosylation of CGTase from various sources. Heliyon 2021, 7, e06305. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.K.; Show, P.L.; Yap, Y.J.; Ariff, A.B.; Mohammad Annuar, M.S.; Lai, O.M.; Tang, T.K.; Juan, J.C.; Ling, T.C. Production of gamma-cyclodextrin by Bacillus cereus cyclodextrin glycosyltransferase using extractive bioconversion in polymer-salt aqueous two-phase system. J. Biosci. Bioeng. 2016, 121, 692–696. [Google Scholar] [CrossRef]
- Leemhuis, H.; Kelly, R.M.; Dijkhuizen, L. Engineering of cyclodextrin glucanotransferases and the impact for biotechnological applications. Appl. Microbiol. Biotechnol. 2010, 85, 823–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, K.; Kashyap, A.; Saini, M.; Gupta, R. Gamma cyclodextrin glycosyltransferase from evansella caseinilytica: Production, characterization and product specificity. 3 Biotech 2022, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Jemli, S.; Jaoua, M.; Bejar, S. US132 Cyclodextrin Glucanotransferase Engineering by Random Mutagenesis for an Anti-Staling Purpose. Mol. Biotechnol. 2016, 58, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Sonnendecker, C.; Zimmermann, W. Domain shuffling of cyclodextrin glucanotransferases for tailored product specificity and thermal stability. FEBS Open Bio 2019, 9, 384–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ottoni, J.R.; e Silva, T.R.; de Oliveira, V.M.; Passarini, M.R.Z. Characterization of amylase produced by cold-adapted bacteria from Antarctic samples. Biocatal. Agric. Biotechnol. 2020, 23, 101452. [Google Scholar] [CrossRef]
- Couto, S.R.; Sanromán, M.Á. Application of solid-state fermentation to food industry—A review. J. Food Eng. 2006, 76, 291–302. [Google Scholar] [CrossRef]
- Tamman, H.; Ainelo, A.; Tagel, M.; Hõrak, R. Stability of the GraA Antitoxin Depends on Growth Phase, ATP Level, and Global Regulator MexT. J. Bacteriol. 2015, 198, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, B.M.; Breidenstein, E.B.M.; de la Fuente-Núñez, C.; Reffuveille, F.; Mawla, G.D.; Hancock, R.E.W.; Baker, T.A. Two Isoforms of Clp Peptidase in Pseudomonas aeruginosa Control Distinct Aspects of Cellular Physiology. J. Bacteriol. 2017, 199, e00568-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Yu, Y.; Qi, Y.; Wang, F.; Yan, J.; Zou, H. Peptide profiling and the bioactivity character of yogurt in the simulated gastrointestinal digestion. J. Proteomics 2016, 141, 24–46. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The Proteome of Native Adult Muller Glial Cells from Murine Retina. Mol. Cell Proteomics 2016, 15, 462–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisniewski, J.R.; Zielinska, D.F.; Mann, M. Comparison of ultrafiltration units for proteomic and N-glycoproteomic analysis by the filter-aided sample preparation method. Anal. Biochem. 2011, 410, 307–309. [Google Scholar] [CrossRef]
- Kleinwort, K.J.H.; Degroote, R.L.; Hirmer, S.; Korbonits, L.; Lorenz, L.; Scholz, A.M.; Hauck, S.M.; Deeg, C.A. Bovine Peripheral Blood Derived Lymphocyte Proteome and Secretome Show Divergent Reaction of Bovine Immune Phenotypes after Stimulation with Pokeweed Mitogen. Proteomes 2022, 10, 7. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleinwort, K.J.H.; Weigand, M.; Hoffmann, L.; Degroote, R.L.; Dietrich, R.; Märtlbauer, E.; Hauck, S.M.; Deeg, C.A. Pudding Proteomics: Cyclomaltodextrin Glucanotransferase and Microbial Proteases Can Liquefy Extended Shelf Life Dairy Products. Metabolites 2022, 12, 254. https://doi.org/10.3390/metabo12030254
Kleinwort KJH, Weigand M, Hoffmann L, Degroote RL, Dietrich R, Märtlbauer E, Hauck SM, Deeg CA. Pudding Proteomics: Cyclomaltodextrin Glucanotransferase and Microbial Proteases Can Liquefy Extended Shelf Life Dairy Products. Metabolites. 2022; 12(3):254. https://doi.org/10.3390/metabo12030254
Chicago/Turabian StyleKleinwort, Kristina J. H., Maria Weigand, Lydia Hoffmann, Roxane L. Degroote, Richard Dietrich, Erwin Märtlbauer, Stefanie M. Hauck, and Cornelia A. Deeg. 2022. "Pudding Proteomics: Cyclomaltodextrin Glucanotransferase and Microbial Proteases Can Liquefy Extended Shelf Life Dairy Products" Metabolites 12, no. 3: 254. https://doi.org/10.3390/metabo12030254
APA StyleKleinwort, K. J. H., Weigand, M., Hoffmann, L., Degroote, R. L., Dietrich, R., Märtlbauer, E., Hauck, S. M., & Deeg, C. A. (2022). Pudding Proteomics: Cyclomaltodextrin Glucanotransferase and Microbial Proteases Can Liquefy Extended Shelf Life Dairy Products. Metabolites, 12(3), 254. https://doi.org/10.3390/metabo12030254