Untargeted Metabolomics Showed Accumulation of One-Carbon Metabolites to Facilitate DNA Methylation during Extracellular Matrix Detachment of Cancer Cells
Abstract
:1. Introduction
2. Results
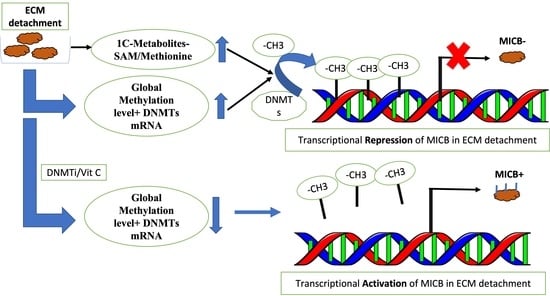
2.1. Loss of ECM Attachment Increased Levels of One-Carbon (1C) Metabolites
2.2. Loss of ECM Attachment Increased Expression of One-Carbon Pathway-Related Metabolic Genes and Global DNA Methylation
2.3. Loss of ECM Attachment Represses Expression of NKG2DLs by Inducing Promoter Methylation
2.4. Vitamin C—A Global Methylation Inhibitor Reduces DNA Methylation and Induces MICA/B Expression during ECM Detachment
3. Discussion
4. Methods
4.1. Cell Culture and Viability Assay
4.2. Metabolites Extraction
4.3. Analysis of HPLC Coupled LC–MS/MS
4.4. Quantitative Real-Time Reverse Transcription PCR
4.5. Quantitative Assay of DNA Methylcytosine (5mC) Level
4.6. Flow Cytometry
4.7. Bisulfite Conversion and Methylation-Specific PCR (MSP)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Buchheit, C.L.; Weigel, K.J.; Schafer, Z.T. Cancer Cell Survival during Detachment from the ECM: Multiple Barriers to Tumour Progression. Nat. Rev. Cancer 2014, 14, 632–641. [Google Scholar] [CrossRef]
- Paoli, P.; Giannoni, E.; Chiarugi, P. Anoikis Molecular Pathways and Its Role in Cancer Progression. Biochim. Et. Biophys. Acta-Mol. Cell Res. 2013, 1833, 3481–3498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mason, J.A.; Davison-Versagli, C.A.; Leliaert, A.K.; Pape, D.J.; McCallister, C.; Zuo, J.; Durbin, S.M.; Buchheit, C.L.; Zhang, S.; Schafer, Z.T. Oncogenic Ras Differentially Regulates Metabolism and Anoikis in Extracellular Matrix-Detached Cells. Cell Death Differ. 2016, 23, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranganathan, S.; Kumar, S.; Mohanty, S.S.; Jolly, M.K.; Rangarajan, A. Cellular Plasticity in Matrix-Attached and -Detached Cells: Implications in Metastasis. J. Indian Inst. Sci. 2020, 100, 525–536. [Google Scholar] [CrossRef]
- Sousa, B.; Pereira, J.; Paredes, J. The Crosstalk between Cell Adhesion and Cancer Metabolism. Int. J. Mol. Sci. 2019, 20, 1933. [Google Scholar] [CrossRef] [Green Version]
- Elia, I.; Doglioni, G.; Fendt, S.M. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018, 28, 673–684. [Google Scholar] [CrossRef]
- Labuschagne, C.F.; Cheung, E.C.; Blagih, J.; Domart, M.C.; Vousden, K.H. Cell Clustering Promotes a Metabolic Switch That Supports Metastatic Colonization. Cell Metab. 2019, 30, 720–734. [Google Scholar] [CrossRef] [Green Version]
- Eales, K.L.; Hollinshead, K.E.R.; Tennant, D.A. Hypoxia and Metabolic Adaptation of Cancer Cells. Oncogenesis 2016, 5, e190. [Google Scholar] [CrossRef] [Green Version]
- Bacigalupa, Z.A.; Rathmell, W.K. Beyond Glycolysis: Hypoxia Signaling as a Master Regulator of Alternative Metabolic Pathways and the Implications in Clear Cell Renal Cell Carcinoma. Cancer Lett. 2020, 489, 19–28. [Google Scholar] [CrossRef]
- Rankin, E.B.; Nam, J.M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Gilkes, D.M.; Semenza, G.L.; Wirtz, D. Hypoxia and the Extracellular Matrix: Drivers of Tumour Metastasis. Nat. Rev. Cancer 2014, 14, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, S.; Zhang, J.; Li, F.; Du, W.; Zhou, X.; Wan, M.; Fan, Y.; Xu, X.; Zhou, X.; Zheng, L.; et al. One-Carbon Metabolism Links Nutrition Intake to Embryonic Development via Epigenetic Mechanisms. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA Methylation: Roles in Mammalian Development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Cahill, C.M.; Huang, X.; Roffman, J.L.; Lamon-Fava, S.; Fava, M.; Mischoulon, D.; Rogers, J.T. S-Adenosyl Methionine and Transmethylation Pathways in Neuropsychiatric Diseases Throughout Life. Neurotherapeutics 2018, 15, 156–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reik, W.; Dean, W.; Walter, J. Epigenetic Reprogramming in Mammalian Development. Science 2001, 293, 1089–1093. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, A.M.; Ali, M.M. Methyl Donor Micronutrients That Modify DNA Methylation and Cancer Outcome. Nutrients 2019, 11, 608. [Google Scholar] [CrossRef] [Green Version]
- Lyko, F. The DNA Methyltransferase Family: A Versatile Toolkit for Epigenetic Regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Shait Mohammed, M.R.; Alghamdi, R.A.; Alzahrani, A.M.; Zamzami, M.A.; Choudhry, H.; Khan, M.I. Compound C, a Broad Kinase Inhibitor Alters Metabolic Fingerprinting of Extra Cellular Matrix Detached Cancer Cells. Front. Oncol. 2021, 11, 1. [Google Scholar] [CrossRef]
- Paul, S.; Lal, G. The Molecular Mechanism of Natural Killer Cells Function and Its Importance in Cancer Immunotherapy. Front. Immunol. 2017, 8, 1. [Google Scholar] [CrossRef] [Green Version]
- Eagle, R.; Jafferji, I.; Barrow, A. Beyond Stressed Self: Evidence for NKG2D Ligand Expression on Healthy Cells. Curr. Immunol. Rev. 2009, 5, 22–34. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, A.M.; Diefenbach, A.; McMahon, C.W.; Xiong, N.; Carlyle, J.R.; Raulet, D.H. The Role of the NKG2D Immunoreceptor in Immune Cell Activation and Natural Killing. Immunity 2002, 17, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Nausch, N.; Cerwenka, A. NKG2D Ligands in Tumor Immunity. Oncogene 2008, 27, 5944–5958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghadially, H.; Brown, L.; Lloyd, C.; Lewis, L.; Lewis, A.; Dillon, J.; Sainson, R.; Jovanovic, J.; Tigue, N.J.; Bannister, D.; et al. MHC Class i Chain-Related Protein A and B (MICA and MICB) Are Predominantly Expressed Intracellularly in Tumour and Normal Tissue. Br. J. Cancer 2017, 116, 1208–1217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zingoni, A.; Molfetta, R.; Fionda, C.; Soriani, A.; Paolini, R.; Cippitelli, M.; Cerboni, C.; Santoni, A. NKG2D and Its Ligands: “One for All, All for One”. Front. Immunol. 2018, 9, 476. [Google Scholar] [CrossRef]
- Bugide, S.; Green, M.R.; Wajapeyee, N. Inhibition of Enhancer of Zeste Homolog 2 (EZH2) Induces Natural Killer Cell-Mediated Eradication of Hepatocellular Carcinoma Cells. Proc. Natl. Acad. Sci. USA 2018, 115, E3509–E3518. [Google Scholar] [CrossRef] [Green Version]
- Kato, N.; Tanaka, J.; Sugita, J.; Toubai, T.; Miura, Y.; Ibata, M.; Syono, Y.; Ota, S.; Kondo, T.; Asaka, M.; et al. Regulation of the Expression of MHC Class I-Related Chain A, B (MICA, MICB) via Chromatin Remodeling and Its Impact on the Susceptibility of Leukemic Cells to the Cytotoxicity of NKG2D-Expressing Cells. Leukemia 2007, 21, 2103–2108. [Google Scholar] [CrossRef]
- Sers, C.; Kuner, R.; Falk, C.S.; Lund, P.; Sueltmann, H.; Braun, M.; Buness, A.; Ruschhaupt, M.; Conrad, J.; Mang-Fatehi, S.; et al. Down-Regulation of HLA Class I and NKG2D Ligands through a Concerted Action of MAPK and DNA Methyltransferases in Colorectal Cancer Cells. Int. J. Cancer 2009, 125, 1626–1639. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’his, D. The Diverse Roles of DNA Methylation in Mammalian Development and Disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Baraganõ Raneros, A.; Martín-Palanco, V.; Fernandez, A.F.; Rodriguez, R.M.; Fraga, M.F.; Lopez-Larrea, C.; Suarez-Alvarez, B. Methylation of NKG2D Ligands Contributes to Immune System Evasion in Acute Myeloid Leukemia. Genes Immun. 2015, 16, 71–82. [Google Scholar] [CrossRef]
- Lee Chong, T.; Ahearn, E.L.; Cimmino, L. Reprogramming the Epigenome with Vitamin C. Front. Cell Dev. Biol. 2019, 7, 128. [Google Scholar] [CrossRef]
- Young, J.I.; Züchner, S.; Wang, G. Regulation of the Epigenome by Vitamin C. Annu. Rev. Nutr. 2015, 35, 545–564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moncayo, G.; Lin, D.; McCarthy, M.T.; Watson, A.A.; O’Callaghan, C.A. MICA Expression Is Regulated by Cell Adhesion and Contact in a FAK/Src-Dependent Manner. Front. Immunol. 2017, 7, 687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thienpont, B.; Steinbacher, J.; Zhao, H.; D’Anna, F.; Kuchnio, A.; Ploumakis, A.; Ghesquière, B.; Van Dyck, L.; Boeckx, B.; Schoonjans, L.; et al. Tumour Hypoxia Causes DNA Hypermethylation by Reducing TET Activity. Nature 2016, 537, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Fouad, M.A.; Salem, S.E.; Hussein, M.M.; Zekri, A.R.N.; Hafez, H.F.; El Desouky, E.D.; Shouman, S.A. Impact of Global DNA Methylation in Treatment Outcome of Colorectal Cancer Patients. Front. Pharmacol. 2018, 9, 1173. [Google Scholar] [CrossRef] [PubMed]
- Guerra, N.; Tan, Y.X.; Joncker, N.T.; Choy, A.; Gallardo, F.; Xiong, N.; Knoblaugh, S.; Cado, D.; Greenberg, N.R.; Raulet, D.H. NKG2D-Deficient Mice Are Defective in Tumor Surveillance in Models of Spontaneous Malignancy. Immunity 2008, 28, 571–580. [Google Scholar] [CrossRef] [Green Version]
- Raulet, D.H.; Gasser, S.; Gowen, B.G.; Deng, W.; Jung, H. Regulation of Ligands for the NKG2D Activating Receptor. Annu. Rev. Immunol. 2013, 31, 413–441. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Huff, T.C.; Sant, D.W.; Camarena, V.; Van Booven, D.; Andrade, N.S.; Mustafi, S.; Monje, P.V.; Wang, G. Vitamin C Regulates Schwann Cell Myelination by Promoting DNA Demethylation of Pro-Myelinating Genes. J. Neurochem. 2020, 157, 1759–1773. [Google Scholar] [CrossRef]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA Methylation Inhibitor 5-Aza-2′-Deoxycytidine Induces Reversible Genome-Wide DNA Damage That Is Distinctly Influenced by DNA Methyltransferases 1 and 3B. Mol. Cell. Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef] [Green Version]
- Mason, J.A.; Hagel, K.R.; Hawk, M.A.; Schafer, Z.T. Metabolism during ECM Detachment: Achilles Heel of Cancer Cells? Trends Cancer 2017, 3, 475–481. [Google Scholar] [CrossRef]
- Endo, H.; Owada, S.; Inagaki, Y.; Shida, Y.; Tatemichi, M. Metabolic Reprogramming Sustains Cancer Cell Survival Following Extracellular Matrix Detachment. Redox Biol. 2020, 36, 101643. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.L.; Xu, X.; Wallenstein, S.; Chen, J. Gene Expression Profiles of the One-Carbon Metabolism Pathway. J. Genet. Genom. 2009, 36, 277–282. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.; Wang, Z.; Zhang, K.; Chi, Z.; Xu, T.; Jiang, D.; Chen, S.; Li, W.; Yang, X.; Zhang, X.; et al. One-Carbon Metabolism Supports S-Adenosylmethionine and Histone Methylation to Drive Inflammatory Macrophages. Mol. Cell 2019, 75, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Mao, Z.; Hwang, J.J.; Lu, S.C. Differential Expression of Methionine Adenosyltransferase Genes Influences the Rate of Growth of Human Hepatocellular Carcinoma Cells. Cancer Res. 1998, 58, 1444–1450. [Google Scholar]
- Newman, A.C.; Maddocks, O.D.K. One-Carbon Metabolism in Cancer. Br. J. Cancer 2017, 116, 1499–1504. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Zhao, J.S.; Li, J.J.; Peng, D.N.; Wang, X.Y.; Chen, T.L.; Qiu, Y.P.; Chen, P.P.; Li, W.J.; Xu, L.Y.; et al. A Combined Proteomics and Metabolomics Profiling of Gastric Cardia Cancer Reveals Characteristic Dysregulations in Glucose Metabolism. Mol. Cell. Proteom. 2010, 9, 2617–2628. [Google Scholar] [CrossRef] [Green Version]
- Gotanda, K.; Hirota, T.; Matsumoto, N.; Ieiri, I. MicroRNA-433 Negatively Regulates the Expression of Thymidylate Synthase (TYMS) Responsible for 5-Fluorouracil Sensitivity in HeLa Cells. BMC Cancer 2013, 13, 369. [Google Scholar] [CrossRef] [Green Version]
- Organista-Nava, J.; Gómez-Gómez, Y.; Del Moral-Hernandez, O.; Illades-Aguiar, B.; Gómez-Santamaria, J.; Rivera-Ramírez, A.B.; Saavedra-Herrera, M.V.; Jimenez-López, M.A.; Leyva-Vázquez, M.A. Deregulation of Folate Pathway Gene Expression Correlates with Poor Prognosis in Acute Leukemia. Oncol. Lett. 2019, 18, 3115–3127. [Google Scholar] [CrossRef]
- Paone, A.; Marani, M.; Fiascarelli, A.; Rinaldo, S.; Giardina, G.; Contestabile, R.; Paiardini, A.; Cutruzzolà, F. SHMT1 Knockdown Induces Apoptosis in Lung Cancer Cells by Causing Uracil Misincorporation. Cell Death Dis. 2014, 5, e1525. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, J. One-Carbon Metabolism and Breast Cancer: An Epidemiological Perspective. J. Genet. Genom. Yi Chuan Xue Bao 2009, 36, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Asai, A.; Konno, M.; Koseki, J.; Taniguchi, M.; Vecchione, A.; Ishii, H. One-Carbon Metabolism for Cancer Diagnostic and Therapeutic Approaches. Cancer Lett. 2020, 470, 141–148. [Google Scholar] [CrossRef] [PubMed]
- van der Wijst, M.G.P.; Venkiteswaran, M.; Chen, H.; Xu, G.L.; Plösch, T.; Rots, M.G. Local Chromatin Microenvironment Determines DNMT Activity: From DNA Methyltransferase to DNA Demethylase or DNA Dehydroxymethylase. Epigenetics 2015, 10, 671–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desjobert, C.; El Maï, M.; Gérard-Hirne, T.; Guianvarćh, D.; Carrier, A.; Pottier, C.; Arimondo, P.B.; Riond, J. Combined Analysis of DNA Methylation and Cell Cycle in Cancer Cells. Epigenetics 2015, 10, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, S.; Sinnberg, T.W.; Berger, A.; Noor, S.; Levesque, M.; Böcker, A.; Niessner, H.; Lauer, U.M.; Bitzer, M.; Garbe, C.; et al. Epigenetic Impacts of Ascorbate on Human Metastatic Melanoma Cells. Front. Oncol. 2014, 4, 227. [Google Scholar] [CrossRef] [Green Version]
- Rocha, M.A.; Veronezi, G.M.B.; Felisbino, M.B.; Gatti, M.S.V.; Tamashiro, W.M.S.C.; Mello, M.L.S. Sodium Valproate and 5-Aza-2′-Deoxycytidine Differentially Modulate DNA Demethylation in G1 Phase-Arrested and Proliferative HeLa Cells. Sci. Rep. 2019, 9, 18236. [Google Scholar] [CrossRef]
- Ritter, C.; Fan, K.; Paulson, K.G.; Nghiem, P.; Schrama, D.; Becker, J.C. Reversal of Epigenetic Silencing of MHC Class i Chain-Related Protein A and B Improves Immune Recognition of Merkel Cell Carcinoma. Sci. Rep. 2016, 6, 21678. [Google Scholar] [CrossRef] [Green Version]
- Sajadian, S.O.; Ehnert, S.; Vakilian, H.; Koutsouraki, E.; Damm, G.; Seehofer, D.; Thasler, W.; Dooley, S.; Baharvand, H.; Sipos, B.; et al. Induction of Active Demethylation and 5hmC Formation by 5-Azacytidine Is TET2 Dependent and Suggests New Treatment Strategies against Hepatocellular Carcinoma. Clin. Epigenetics 2015, 7, 98. [Google Scholar] [CrossRef] [Green Version]
- AlGhamdi, A.A.; Mohammed, M.R.S.; Zamzami, M.A.; Al-Malki, A.L.; Qari, M.H.; Khan, M.I.; Choudhry, H. Untargeted Metabolomics Identifies Key Metabolic Pathways Altered by Thymoquinone in Leukemic Cancer Cells. Nutrients 2020, 12, 1792. [Google Scholar] [CrossRef]
- Wang, X.; Guo, X.; Yu, W.; Li, C.; Gui, Y.; Cai, Z. Expression of Methionine Adenosyltransferase 2A in Renal Cell Carcinomas and Potential Mechanism for Kidney Carcinogenesis. BMC Cancer 2014, 14, 196. [Google Scholar] [CrossRef] [Green Version]
- Avila, M.A.; Berasain, C.; Torres, L.; Martín-Duce, A.; Corrales, F.J.; Yang, H.; Prieto, J.; Lu, S.C.; Caballería, J.; Rodés, J.; et al. Reduced MRNA Abundance of the Main Enzymes Involved in Methionine Metabolism in Human Liver Cirrhosis and Hepatocellular Carcinoma. J. Hepatol. 2000, 33, 907–914. [Google Scholar] [CrossRef] [Green Version]
- Bing, Y.; Zhu, S.; Yu, G.; Li, T.; Liu, W.; Li, C.; Wang, Y.; Qi, H.; Guo, T.; Yuan, Y.; et al. Glucocorticoid-Induced S-Adenosylmethionine Enhances the Interferon Signaling Pathway by Restoring STAT1 Protein Methylation in Hepatitis B Virus-Infected Cells. J. Biol. Chem. 2014, 289, 32639–32655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakami, K.; Ooyama, A.; Ruszkiewicz, A.; Jin, M.; Watanabe, G.; Moore, J.; Oka, T.; Iacopetta, B.; Minamoto, T. Low Expression of γ-Glutamyl Hydrolase MRNA in Primary Colorectal Cancer with the CpG Island Methylator Phenotype. Br. J. Cancer 2008, 98, 1555–1561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| MICA | CCT GCA ATC CCA GCA CTT TG | ATT CAC CAC CAA GCC CGT CT |
| MICB | CAC GTT CGC CCT TTG TTC AG | GGA GGC AGA GGT TGC AGT GA |
| DNMT1 | CAG CAA CGG GCA GAT GTT TC | CGG AGG GGTG CTTTGT AGA TG |
| DNMT3a | CTA CGC ACC ACC TCC ACC AG | CAA TGT TCC GGC ACT TCT GC |
| DNMT3b | GAG TCC ATT GCT GTT GGA ACC G | ATG TCC CTC TTG TCG CCA ACC T |
| MAT2A | ATGAACGGACAGCTCAACGG | CCAGCAAGAAGGATCATTCCAG |
| MAT2A | ATGAACGGACAGCTCAACGG | CCAGCAAGAAGGATCATTCCAG |
| AHCY | GCTGGAAGTTGGAGTTCTCGC | GTCCTCCCGCTGCTGTCA |
| BHMT | GTC ATG CAG ACC TTC ACC TTC TA | CTC CTT CAT GAG CTT CAC TG |
| CBS | ACA TGA CCA AGT TCC TGA GC | GCC ACG AAG TTC AGC AAG TC |
| DHFR | GTCCTCCCGCTGCTGTCA | GCCGATGCCCATGTTCTG |
| MTHFD1 | CGTGGGCAGCGGACTAA | CCTTATTTGCGCGGAGATCT |
| MTHFR | GGCCATCTGCACAAAGCTAAG | AACTCACTTCGGATGTGCTTCAC |
| TYMS | TCTGGAAGGGTGTTTTGGAG | CCTCCACTGGAAGCCATAAA |
| SHMT1 | AGGAAAGGAGTGAAAAGTGTGGAT | GACACCAGTGTCGCTCTGGATCTG |
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| MICA methylated | TTA TTG TTA GTA ACG TTG TGC GC | AAC CTA AAA CAA AAA CCA ACT TCG |
| MICA unmethylated | TTA TTG TTA GTA ATG TTG TGT GTG G | ACC TAA AAC AAA AAC CAA CTT CAA A |
| MICB methylated | GTT GGG ATT ATA GAG GTG AGT TAT C | CTC AAA AAA ACT AAT TTA TCC GAA |
| MICB unmethylated | TGT TGG GAT TAT AGA GGT GAG TTA TT | CCT CAA AAA AAC TAA TTT ATC CAA A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nur, S.M.; Shait Mohammed, M.R.; Zamzami, M.A.; Choudhry, H.; Ahmad, A.; Ateeq, B.; Rather, I.A.; Khan, M.I. Untargeted Metabolomics Showed Accumulation of One-Carbon Metabolites to Facilitate DNA Methylation during Extracellular Matrix Detachment of Cancer Cells. Metabolites 2022, 12, 267. https://doi.org/10.3390/metabo12030267
Nur SM, Shait Mohammed MR, Zamzami MA, Choudhry H, Ahmad A, Ateeq B, Rather IA, Khan MI. Untargeted Metabolomics Showed Accumulation of One-Carbon Metabolites to Facilitate DNA Methylation during Extracellular Matrix Detachment of Cancer Cells. Metabolites. 2022; 12(3):267. https://doi.org/10.3390/metabo12030267
Chicago/Turabian StyleNur, Suza Mohammad, Mohammed Razeeth Shait Mohammed, Mazin A. Zamzami, Hani Choudhry, Aamir Ahmad, Bushra Ateeq, Irfan A. Rather, and Mohammad Imran Khan. 2022. "Untargeted Metabolomics Showed Accumulation of One-Carbon Metabolites to Facilitate DNA Methylation during Extracellular Matrix Detachment of Cancer Cells" Metabolites 12, no. 3: 267. https://doi.org/10.3390/metabo12030267
APA StyleNur, S. M., Shait Mohammed, M. R., Zamzami, M. A., Choudhry, H., Ahmad, A., Ateeq, B., Rather, I. A., & Khan, M. I. (2022). Untargeted Metabolomics Showed Accumulation of One-Carbon Metabolites to Facilitate DNA Methylation during Extracellular Matrix Detachment of Cancer Cells. Metabolites, 12(3), 267. https://doi.org/10.3390/metabo12030267