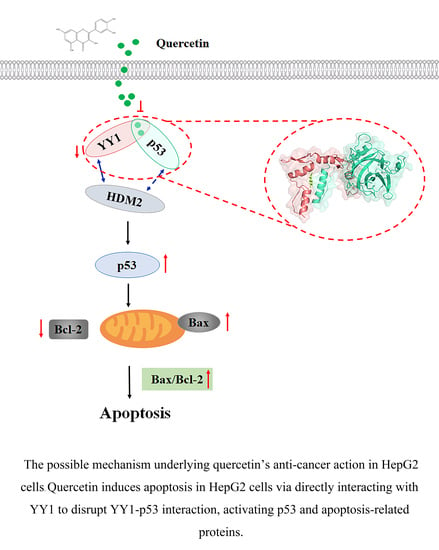
Quercetin Induces Apoptosis in HepG2 Cells via Directly Interacting with YY1 to Disrupt YY1-p53 Interaction
Abstract
:1. Introduction
2. Material and Methods
2.1. Materials
2.2. Cell Culture
2.3. Cell Viability Assays
2.4. Transfection Experiments
2.5. Apoptosis Assays
2.6. Western Blot Experiments
2.7. Molecular Docking
2.8. Preparation of YY1 Protein and Quercetin Solutions
2.9. The Cellular Thermal Shift Assay (CETSA)
2.10. UV-Vis Absorption Spectroscopy for Examining YY1-Quercetin Mixtures
2.11. YY1-Quercetin Interaction Revealed by Fluorescence Spectroscopy
2.12. YY1-Quercetin Interaction Revealed by Circular Dichroism (CD)
2.13. Statistical Analysis
3. Results
3.1. Quercetin Inhibited Cell Viability in HepG2 Cells
3.2. Quercetin Induced Apoptosis by Regulating p53-Mediated Signaling Pathway in HepG2 Cells
3.3. YY1 and Its Role in Quercetin-Induced Apoptosis of HepG2 Cells
3.4. The Interaction between Quercetin and YY1
3.4.1. Molecular Docking Showed the Interactions between Quercetin and YY1
3.4.2. The Direct Binding of Quercetin to YY1 Revealed by CETSA
3.4.3. UV–Vis Absorption Spectra of Quercetin–YY1 Systems
3.4.4. Fluorescence Spectroscopy of YY1 Induced by Quercetin
3.4.5. Quercetin-Induced Changes in the Conformation of YY1 Protein Revealed by Circular Dichroism (CD) Spectroscopy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Huang, C.; Wei, Y.X.; Shen, M.C.; Tu, Y.H.; Wang, C.C.; Huang, H.C. Chrysin, Abundant in Morinda citrifolia Fruit Water-EtOAc Extracts, Combined with Apigenin Synergistically Induced Apoptosis and Inhibited Migration in Human Breast and Liver Cancer Cells. J. Agric. Food Chem. 2016, 64, 4235–4245. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Liu, K.; Shen, Q.; Li, Q.; Hao, J.; Han, F.; Jiang, R.W. Reversal of Multidrug Resistance in Cancer by Multi-Functional Flavonoids. Front. Oncol. 2019, 9, 487. [Google Scholar] [CrossRef]
- Anand, J.; Rai, N. Anticandidal synergistic activity of green tea catechins, antimycotics and copper sulphate as a mean of combinational drug therapy against candidiasis. J. Mycol. Med. 2017, 27, 33–45. [Google Scholar] [CrossRef]
- Almatroodi, S.A.; Alsahli, M.A.; Almatroudi, A.; Verma, A.K.; Aloliqi, A.; Allemailem, K.S.; Khan, A.A.; Rahmani, A.H. Potential Therapeutic Targets of Quercetin, a Plant Flavonol, and Its Role in the Therapy of Various Types of Cancer through the Modulation of Various Cell Signaling Pathways. Molecules 2021, 26, 1315. [Google Scholar] [CrossRef] [PubMed]
- Abotaleb, M.; Samuel, S.M.; Varghese, E.; Varghese, S.; Kubatka, P.; Liskova, A.; Büsselberg, D. Flavonoids in Cancer and Apoptosis. Cancers 2018, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Kopustinskiene, D.M.; Jakstas, V.; Savickas, A.; Bernatoniene, J. Flavonoids as Anticancer Agents. Nutrients 2020, 12, 457. [Google Scholar] [CrossRef]
- Maurya, A.K.; Vinayak, M. Quercetin regresses Dalton’s lymphoma growth via suppression of PI3K/AKT signaling leading to upregulation of p53 and decrease in energy metabolism. Nutr. Cancer 2015, 67, 354–363. [Google Scholar] [CrossRef]
- He, D.; Guo, X.; Zhang, E.; Zi, F.; Chen, J.; Chen, Q.; Lin, X.; Yang, L.; Li, Y.; Wu, W.; et al. Quercetin induces cell apoptosis of myeloma and displays a synergistic effect with dexamethasone in vitro and in vivo xenograft models. Oncotarget 2016, 7, 45489–45499. [Google Scholar] [CrossRef]
- Dong, Y.; Yang, J.; Yang, L.; Li, P. Quercetin Inhibits the Proliferation and Metastasis of Human Non-Small Cell Lung Cancer Cell Line: The Key Role of Src-Mediated Fibroblast Growth Factor-Inducible 14 (Fn14)/Nuclear Factor kappa B (NF-kappaB) pathway. Med. Sci. Monit. 2020, 26, e920537. [Google Scholar] [CrossRef]
- Gao, X.; Wang, B.; Wei, X.; Men, K.; Zheng, F.; Zhou, Y.; Zheng, Y.; Gou, M.; Huang, M.; Guo, G.; et al. Anticancer effect and mechanism of polymer micelle-encapsulated quercetin on ovarian cancer. Nanoscale 2012, 4, 7021–7030. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, Y.; Liao, J.; Gong, C.; Sun, C.; Zhou, X.; Wei, X.; Zhang, T.; Gao, Q.; Ma, D.; et al. Quercetin induces endoplasmic reticulum stress to enhance cDDP cytotoxicity in ovarian cancer: Involvement of STAT3 signaling. FEBS J. 2015, 282, 1111–1125. [Google Scholar] [CrossRef]
- Zhao, P.; Mao, J.M.; Zhang, S.Y.; Zhou, Z.Q.; Tan, Y.; Zhang, Y. Quercetin induces HepG2 cell apoptosis by inhibiting fatty acid biosynthesis. Oncol. Lett. 2014, 8, 765–769. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Somasagara, R.R.; Hegde, M.; Nishana, M.; Tadi, S.K.; Srivastava, M.; Choudhary, B.; Raghavan, S.C. Quercetin, a Natural Flavonoid Interacts with DNA, Arrests Cell Cycle and Causes Tumor Regression by Activating Mitochondrial Pathway of Apoptosis. Sci. Rep. 2016, 6, 24049. [Google Scholar] [CrossRef]
- Wu, L.; Li, J.; Liu, T.; Li, S.; Feng, J.; Yu, Q.; Zhang, J.; Chen, J.; Zhou, Y.; Ji, J.; et al. Quercetin shows anti-tumor effect in hepatocellular carcinoma LM3 cells by abrogating JAK2/STAT3 signaling pathway. Cancer Med. 2019, 8, 4806–4820. [Google Scholar] [CrossRef]
- Granado-Serrano, A.B.; Martín, M.A.; Bravo, L.; Goya, L.; Ramos, S. Quercetin induces apoptosis via caspase activation, regulation of Bcl-2, and inhibition of PI-3-kinase/Akt and ERK pathways in a human hepatoma cell line (HepG2). J. Nutr. 2006, 136, 2715–2721. [Google Scholar] [CrossRef] [PubMed]
- Hisaka, T.; Sakai, H.; Sato, T.; Goto, Y.; Nomura, Y.; Fukutomi, S.; Fujita, F.; Mizobe, T.; Nakashima, O.; Tanigawa, M.; et al. Quercetin Suppresses Proliferation of Liver Cancer Cell Lines In Vitro. Anticancer Res. 2020, 40, 4695–4700. [Google Scholar] [CrossRef] [PubMed]
- Fernald, K.; Kurokawa, M. Evading apoptosis in cancer. Trends Cell Biol. 2013, 23, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]
- Fridman, J.S.; Lowe, S.W. Control of apoptosis by p53. Oncogene 2003, 22, 9030–9040. [Google Scholar] [CrossRef] [Green Version]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Sui, G.; Affar el, B.; Shi, Y.; Brignone, C.; Wall, N.R.; Yin, P.; Donohoe, M.; Luke, M.P.; Calvo, D.; Grossman, S.R.; et al. Yin Yang 1 is a negative regulator of p53. Cell 2004, 117, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Verheul, T.C.J.; van Hijfte, L.; Perenthaler, E.; Barakat, T.S. The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1. Front. Cell Dev. Biol. 2020, 8, 592164. [Google Scholar] [CrossRef]
- Sarvagalla, S.; Kolapalli, S.P.; Vallabhapurapu, S. The Two Sides of YY1 in Cancer: A Friend and a Foe. Front. Oncol. 2019, 9, 1230. [Google Scholar] [CrossRef] [PubMed]
- Houbaviy, H.B.; Usheva, A.; Shenk, T.; Burley, S.K. Cocrystal structure of YY1 bound to the adeno-associated virus P5 initiator. Proc. Natl. Acad. Sci. USA 1996, 93, 13577–13582. [Google Scholar] [CrossRef]
- Wang, D.; Jiang, Y.; Sun-Waterhouse, D.-X.; Zhai, H.; Guan, H.; Rong, X.; Li, F.; Yu, J.-C.; Li, D.-P. MicroRNA-based regulatory mechanisms underlying the synergistic antioxidant action of quercetin and catechin in H2O2-stimulated HepG2 cells: Roles of BACH1 in Nrf2-dependent pathways. Free. Radic. Biol. Med. 2020, 153, 122–131. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Albrecht, C.; Joseph, R. Lakowicz: Principles of Fluorescence Spectroscopy, 3rd Edition. Anal. Bioanal. Chem. 2008, 390, 1223–1224. [Google Scholar] [CrossRef]
- Liu, Z.J.; Xu, W.; Han, J.; Liu, Q.Y.; Gao, L.F.; Wang, X.H.; Li, X.L. Quercetin induces apoptosis and enhances gemcitabine therapeutic efficacy against gemcitabine-resistant cancer cells. Anticancer Drugs 2020, 31, 684–692. [Google Scholar] [CrossRef]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef] [Green Version]
- Huttemann, M.; Pecina, P.; Rainbolt, M.; Sanderson, T.H.; Kagan, V.E.; Samavati, L.; Doan, J.W.; Lee, I. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion 2011, 11, 369–381. [Google Scholar] [CrossRef]
- Teng, Y.; Ji, F.; Li, C.; Yu, Z.; Liu, R. Interaction mechanism between 4-aminoantipyrine and the enzyme lysozyme. J. Lumin. 2011, 131, 2661–2667. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, P.; Yang, Z.; Du, F.; Li, Z.; Wu, C.; Fang, A.; Xu, X.; Zhou, G. Molecular dynamics simulation exploration of the interaction between curcumin and myosin combined with the results of spectroscopy techniques. Food Hydrocoll. 2019, 101, 105455. [Google Scholar] [CrossRef]
- Ware, W.R. Oxygen quenching of fluorescence in solution: An experimental study of the diffusion process. J. Phys. Chem. 1962, 66, 455–458. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. A probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-J.; Wang, Y.; Ou-Yang, Y.; Zhou, J.; Liu, Y. Characterize the interaction between naringenin and bovine serum albumin using spectroscopic approach. J. Lumin. 2010, 130, 1394–1399. [Google Scholar] [CrossRef]
- Lambert, J.D.; Yang, C.S. Cancer chemopreventive activity and bioavailability of tea and tea polyphenols. Mutat. Res. 2003, 523–524, 201–208. [Google Scholar] [CrossRef]
- Wang, W.; Sun, C.; Mao, L.; Ma, P.; Liu, F.; Yang, J.; Gao, Y. The biological activities, chemical stability, metabolism and delivery systems of quercetin: A review. Trends Food Sci. Technol. 2016, 56, 21–38. [Google Scholar] [CrossRef]
- Shanmugam, H.; Rengaraja, C.; Natara, S.; Sharma, A. Interactions of plant food bioactives-loaded nano delivery systems at the nano-bio interface and its pharmacokinetics: An overview. Food Front. 2022, 3, 256–275. [Google Scholar] [CrossRef]
- Gang, W.; Jie, W.J.; Ping, Z.L.; Ming, D.S.; Ying, L.J.; Lei, W.; Fang, Y. Liposomal quercetin: Evaluating drug delivery in vitro and biodistribution in vivo. Expert Opin. Drug Deliv. 2012, 9, 599–613. [Google Scholar] [CrossRef]
- Lee, T.; Chang, Y.H. Structural, physicochemical, and in-vitro release properties of hydrogel beads produced by oligochitosan and de-esterified pectin from yuzu (Citrus junos) peel as a quercetin delivery system for colon target. Food Hydrocoll. 2020, 108, 106086. [Google Scholar] [CrossRef]
- Xiong, Q.; Wang, Y.; Wan, J.; Yuan, P.; Chen, H.; Zhang, L. Facile preparation of hyaluronic acid-based quercetin nanoformulation for targeted tumor therapy. Int. J. Biol. Macromol. 2020, 147, 937–945. [Google Scholar] [CrossRef]
- Sadhukhan, P.; Kundu, M.; Chatterjee, S.; Ghosh, N.; Manna, P.; Das, J.; Sil, P.C. Targeted delivery of quercetin via pH-responsive zinc oxide nanoparticles for breast cancer therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 100, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Meulmeester, E.; Jochemsen, A.G. p53: A guide to apoptosis. Curr. Cancer Drug Targets 2008, 8, 87–97. [Google Scholar] [CrossRef]
- Ramos, S. Effects of dietary flavonoids on apoptotic pathways related to cancer chemoprevention. J. Nutr. Biochem. 2007, 18, 427–442. [Google Scholar] [CrossRef]
- Gordon, S.; Akopyan, G.; Garban, H.; Bonavida, B. Transcription factor YY1: Structure, function, and therapeutic implications in cancer biology. Oncogene 2006, 25, 1125–1142. [Google Scholar] [CrossRef]
- Kurisaki, K.; Kurisaki, A.; Valcourt, U.; Terentiev, A.A.; Pardali, K.; Ten Dijke, P.; Heldin, C.H.; Ericsson, J.; Moustakas, A. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 2003, 23, 4494–4510. [Google Scholar] [CrossRef]
- Yakovleva, T.; Kolesnikova, L.; Vukojevic, V.; Gileva, I.; Tan-No, K.; Austen, M.; Luscher, B.; Ekstrom, T.J.; Terenius, L.; Bakalkin, G. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem. Biophys. Res. Commun. 2004, 318, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Zhang, J.; Xiao, W.; Zheng, X. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014, 42, 1799–1811. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Yuan, Y.; Zhang, W.; Guan, W.; Wu, Z.; Jin, C.; Chen, H.; Zhang, L.; Yang, X.; et al. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010, 38, 6544–6554. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, D.; Nguyen, L.X.; Wu, H.; Li, L.; Dong, D.; Troadec, E.; Zhu, Y.; Hoang, D.H.; Stein, A.S.; et al. Targeting cell membrane HDM2: A novel therapeutic approach for acute myeloid leukemia. Leukemia 2020, 34, 75–86. [Google Scholar] [CrossRef]
- Zhou, S.; Li, P.; Qin, L.; Huang, S.; Dang, N. Transcription factor YY1 contributes to human melanoma cell growth through modulating the p53 signalling pathway. Exp. Dermatol. 2022, 31, 1563–1578. [Google Scholar] [CrossRef]
- Yu, Q.; Fan, L.; Duan, Z. Five individual polyphenols as tyrosinase inhibitors: Inhibitory activity, synergistic effect, action mechanism, and molecular docking. Food Chem. 2019, 297, 124910. [Google Scholar] [CrossRef]
- Real Hernandez, L.M.; Fan, J.; Johnson, M.H.; Gonzalez de Mejia, E. Berry Phenolic Compounds Increase Expression of Hepatocyte Nuclear Factor-1alpha (HNF-1alpha) in Caco-2 and Normal Colon Cells Due to High Affinities with Transcription and Dimerization Domains of HNF-1alpha. PLoS ONE 2015, 10, e0138768. [Google Scholar] [CrossRef]
- Li, M.; Huang, W.; Jie, F.; Wang, M.; Zhong, Y.; Chen, Q.; Lu, B. Discovery of Keap1−Nrf2 small−molecule inhibitors from phytochemicals based on molecular docking. Food Chem. Toxicol. 2019, 133, 110758–110766. [Google Scholar] [CrossRef]
- Ramyaa, P.; krishnaswamy, R.; Padma, V.V. Quercetin modulates OTA-induced oxidative stress and redox signalling in HepG2 cells—Up regulation of Nrf2 expression and down regulation of NF-κB and COX-2. Biochim. Biophys. Acta 2014, 1840, 681–692. [Google Scholar] [CrossRef]
- Li, T.; Li, F.; Liu, X.; Liu, J.; Li, D. Synergistic anti-inflammatory effects of quercetin and catechin via inhibiting activation of TLR4-MyD88-mediated NF-kappaB and MAPK signaling pathways. Phytother. Res. 2019, 33, 756–767. [Google Scholar] [CrossRef]
- El-Far, A.H.; Lebda, M.A.; Noreldin, A.E.; Atta, M.S.; Elewa, Y.H.A.; Elfeky, M.; Mousa, S.A. Quercetin Attenuates Pancreatic and Renal D-Galactose-Induced Aging-Related Oxidative Alterations in Rats. Int. J. Mol. Sci. 2020, 21, 4348. [Google Scholar] [CrossRef]
- Liu, W.; Guo, Q.; Zhao, H. Oxidative stress-elicited YY1 potentiates antioxidative response via enhancement of NRF2-driven transcriptional activity: A potential neuronal defensive mechanism against ischemia/reperfusion cerebral injury. Biomed. Pharm. 2018, 108, 698–706. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guan, H.; Zhang, W.; Liu, H.; Jiang, Y.; Li, F.; Wu, M.; Waterhouse, G.I.N.; Sun-Waterhouse, D.; Li, D. Quercetin Induces Apoptosis in HepG2 Cells via Directly Interacting with YY1 to Disrupt YY1-p53 Interaction. Metabolites 2023, 13, 229. https://doi.org/10.3390/metabo13020229
Guan H, Zhang W, Liu H, Jiang Y, Li F, Wu M, Waterhouse GIN, Sun-Waterhouse D, Li D. Quercetin Induces Apoptosis in HepG2 Cells via Directly Interacting with YY1 to Disrupt YY1-p53 Interaction. Metabolites. 2023; 13(2):229. https://doi.org/10.3390/metabo13020229
Chicago/Turabian StyleGuan, Hui, Wenyuan Zhang, Hui Liu, Yang Jiang, Feng Li, Maoyu Wu, Geoffrey I. N. Waterhouse, Dongxiao Sun-Waterhouse, and Dapeng Li. 2023. "Quercetin Induces Apoptosis in HepG2 Cells via Directly Interacting with YY1 to Disrupt YY1-p53 Interaction" Metabolites 13, no. 2: 229. https://doi.org/10.3390/metabo13020229
APA StyleGuan, H., Zhang, W., Liu, H., Jiang, Y., Li, F., Wu, M., Waterhouse, G. I. N., Sun-Waterhouse, D., & Li, D. (2023). Quercetin Induces Apoptosis in HepG2 Cells via Directly Interacting with YY1 to Disrupt YY1-p53 Interaction. Metabolites, 13(2), 229. https://doi.org/10.3390/metabo13020229