A Case Study of Dysfunctional Nicotinamide Metabolism in a 20-Year-Old Male
Abstract
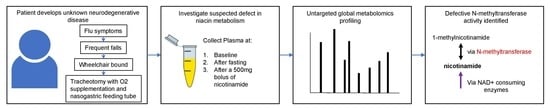
:1. Introduction
2. Case Study
3. Experimental Design
3.1. Sample Collection
3.2. Sample Processing
3.3. Ultrahigh Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC/MS-MS)
3.4. Compound Identification and Data Analysis
4. Results
4.1. Confirmation of Analytical Precision
4.2. Nicotinamide Metabolism
4.3. Methionine Metabolism
4.4. NAD(H) Production
4.5. Inflammatory Response
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ying, W. NAD+/NADH and NADP+/NADPH in Cellular Functions and Cell Death: Regulation and Biological Consequences. Antioxid. Redox Signal 2008, 10, 179–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambon, P.; Weill, J.; Mandel, P. Nicotinamide mononucleotide activation of a new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem. Biophys. Res. Commun. 1963, 11, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.A. Characterization of Five Human cDNAs with Homology to the Yeast SIR2 Gene: Sir2-like Proteins (Sirtuins) Metabolize NAD and May Have Protein ADP-Ribosyltransferase Activity. Biochem. Biophys. Res. Commun. 1999, 260, 273–279. [Google Scholar] [CrossRef]
- Imai, S.-I.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef] [PubMed]
- Landry, J.; Sutton, A.; Tafrov, S.T.; Heller, R.C.; Stebbins, J.; Pillus, L.; Sternglanz, R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 2000, 97, 5807–5811. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.-H.; Yu, X. Human DNA ligase IV is able to use NAD+ as an alternative adenylation donor for DNA ends ligation. Nucleic Acids Res. 2018, 47, 1321–1334. [Google Scholar] [CrossRef] [Green Version]
- Bird, J.G.; Zhang, Y.; Tian, Y.; Panova, N.; Barvík, I.; Greene, L.; Liu, M.; Buckley, B.; Krásný, L.; Lee, J.K.; et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 2016, 535, 444–447. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, J.; Baur, J.A.; Imai, S.-I. NAD+ Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018, 27, 513–528. [Google Scholar] [CrossRef] [Green Version]
- Clement, J.; Wong, M.; Poljak, A.; Sachdev, P.; Braidy, N. The Plasma NAD+ Metabolome Is Dysregulated in “Normal” Aging. Rejuvenation Res. 2019, 22, 121–130. [Google Scholar] [CrossRef] [Green Version]
- Fan, L.; Cacicedo, J.M.; Ido, Y.; Lan, F. Impaired nicotinamide adenine dinucleotide (NAD+) metabolism in diabetes and diabetic tissues: Implications for nicotinamide-related compound treatment. J. Diabetes Investig. 2020, 11, 1403–1419. [Google Scholar] [CrossRef]
- Zhou, C.-C.; Yang, X.; Hua, X.; Liu, J.; Fan, M.-B.; Li, G.-Q.; Song, J.; Xu, T.-Y.; Li, Z.-Y.; Guan, Y.-F.; et al. Hepatic NAD+deficiency as a therapeutic target for non-alcoholic fatty liver disease in ageing. Br. J. Pharmacol. 2016, 173, 2352–2368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; He, H.-J.; Xiong, X.; Zhou, S.; Wang, W.-W.; Feng, L.; Han, R.; Xie, C.-L. NAD+ in Alzheimer’s Disease: Molecular Mechanisms and Systematic Therapeutic Evidence Obtained in vivo. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Abdellatif, M.; Bugger, H.; Kroemer, G.; Sedej, S. NAD+ and Vascular Dysfunction: From Mechanisms to Therapeutic Opportunities. J. Lipid Atheroscler. 2022, 11, 111–132. [Google Scholar] [CrossRef]
- Hu, L.; Ibrahim, K.; Stucki, M.; Frapolli, M.; Shahbeck, N.; Chaudhry, F.A.; Görg, B.; Häussinger, D.; Penberthy, W.T.; Ben-Omran, T.; et al. Secondary NAD+ deficiency in the inherited defect of glutamine synthetase. J. Inherit. Metab. Dis. 2015, 38, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Pirinen, E.; Auranen, M.; Khan, N.A.; Brilhante, V.; Urho, N.; Pessia, A.; Hakkarainen, A.; Kuula, J.; Heinonen, U.; Schmidt, M.S.; et al. Niacin Cures Systemic NAD+ Deficiency and Improves Muscle Performance in Adult-Onset Mitochondrial Myopathy. Cell Metab. 2020, 31, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Braidy, N.; Berg, J.; Clement, J.; Khorshidi, F.; Poljak, A.; Jayasena, T.; Grant, R.; Sachdev, P. Role of Nicotinamide Adenine Dinucleotide and Related Precursors as Therapeutic Targets for Age-Related Degenerative Diseases: Rationale, Biochemistry, Pharmacokinetics, and Outcomes. Antioxid. Redox Signal 2019, 30, 251–294. [Google Scholar] [CrossRef] [PubMed]
- Fessel, J.P.; Oldham, W.M. Nicotine Adenine Dinucleotides: The Redox Currency of the Cell. Antioxid. Redox Signal 2018, 28, 165–166. [Google Scholar] [CrossRef]
- Beaudry, P.; Cohen, P.; Brandel, J.; Delasnerie-Lauprêtre, N.; Richard, S.; Launay, J.; Laplanche, J. 14-3-3 protein, neuron-specific enolase, and S-100 protein in cerebrospinal fluid of patients with Creutzfeldt-Jakob disease. Dement. Geriatr. Cogn. Disord. 1999, 10, 40–46. [Google Scholar] [CrossRef]
- Hsich, G.; Kenney, K.; Gibbs, C.J.; Lee, K.H.; Harrington, M.G. The 14-3-3 Brain Protein in Cerebrospinal Fluid as a Marker for Transmissible Spongiform Encephalopathies. N. Engl. J. Med. 1996, 335, 924–930. [Google Scholar] [CrossRef]
- Jain, M.; Kennedy, A.D.; Elsea, S.; Miller, M.J. Analytes related to erythrocyte metabolism are reliable biomarkers for preanalytical error due to delayed plasma processing in metabolomics studies. Clin. Chim. Acta 2017, 466, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Evans, A.M.; Bridgewater, B.; Liu, Q.; Mitchell, M.; Robinson, R.; Dai, H.; Stewart, S.; Dehaven, C.; Miller, L. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. Metabolomics 2014, 4, 1000132. [Google Scholar] [CrossRef] [Green Version]
- Evans, A.M.; DeHaven, C.D.; Barrett, T.; Mitchell, M.; Milgram, E. Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems. Anal. Chem. 2009, 81, 6656–6667. [Google Scholar] [CrossRef] [PubMed]
- Ford, L.; Kennedy, A.D.; Goodman, K.D.; Pappan, K.L.; Evans, A.M.; Miller, L.A.D.; Wulff, J.E.; Wiggs, B.R.; Lennon, J.J.; Elsea, S.; et al. Precision of a Clinical Metabolomics Profiling Platform for Use in the Identification of Inborn Errors of Metabolism. J. Appl. Lab. Med. 2020, 5, 342–356. [Google Scholar] [CrossRef]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.-M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, A.D.; Wittmann, B.M.; Evans, A.M.; Miller, L.A.; Toal, D.R.; Lonergan, S.; Elsea, S.H.; Pappan, K.L. Metabolomics in the clinic: A review of the shared and unique features of untargeted metabolomics for clinical research and clinical testing. J. Mass Spectrom. 2018, 53, 1143–1154. [Google Scholar] [CrossRef] [PubMed]
- Ford, L.; Mitchell, M.; Wulff, J.; Evans, A.; Kennedy, A.; Elsea, S.; Wittmann, B.; Toal, D. Clinical metabolomics for inborn errors of metabolism. Adv. Clin. Chem. 2022, 107, 79–138. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- Corporation, O. 2022. Available online: https://omicsoftdocs.github.io/ArraySuiteDoc/tutorials/ArrayStudio/ArrayStudio/ (accessed on 30 January 2023).
- Ström, K.; Morales-Alamo, D.; Ottosson, F.; Edlund, A.; Hjort, L.; Jörgensen, S.W.; Almgren, P.; Zhou, Y.; Martin-Rincon, M.; Ekman, C.; et al. N1-methylnicotinamide is a signalling molecule produced in skeletal muscle coordinating energy metabolism. Sci. Rep. 2018, 8, 3016. [Google Scholar] [CrossRef] [Green Version]
- Chlopicki, S.; Swies, J.; Mogielnicki, A.; Buczko, W.; Bartus, M.; Lomnicka, M.; Adamus, J.; Gebicki, J. 1-Methylnicotinamide (MNA), a primary metabolite of nicotinamide, exerts anti-thrombotic activity mediated by a cyclooxygenase-2/prostacyclin pathway. Br. J. Pharmacol. 2007, 152, 230–239. [Google Scholar] [CrossRef]
- Kilgour, M.K.; MacPherson, S.; Zacharias, L.G.; Ellis, A.E.; Sheldon, R.D.; Liu, E.Y.; Keyes, S.; Pauly, B.; Carleton, G.; Allard, B.; et al. 1-Methylnicotinamide is an immune regulatory metabolite in human ovarian cancer. Sci. Adv. 2021, 7. [Google Scholar] [CrossRef]
- Schmitz, G.; Ecker, J. The opposing effects of n−3 and n−6 fatty acids. Prog. Lipid Res. 2008, 47, 147–155. [Google Scholar] [CrossRef]
- Bagga, D.; Wang, L.; Farias-Eisner, R.; Glaspy, J.A.; Reddy, S.T. Differential effects of prostaglandin derived from ω-6 and ω-3 polyunsaturated fatty acids on COX-2 expression and IL-6 secretion. Proc. Natl. Acad. Sci. USA 2003, 100, 1751–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.G.; Stone, N.J. Antiatherosclerotic and Antithrombotic Effects of Omega-3 Fatty Acids. Am. J. Cardiol. 2006, 98, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Martin, N.I.; van Haren, M.J. Nicotinamide N-methyl transferase (NNMT): An emerging therapeutic target. Drug Discov. Today 2021, 26, 2699–2706. [Google Scholar] [CrossRef]
- Roberti, A.; Fernández, A.F.; Fraga, M.F. Nicotinamide N-methyltransferase: At the crossroads between cellular metabolism and epigenetic regulation. Mol. Metab. 2021, 45, 101165. [Google Scholar] [CrossRef]
- Liu, J.-R.; Deng, Z.-H.; Zhu, X.-J.; Zeng, Y.-R.; Guan, X.-X.; Li, J.-H. Roles of Nicotinamide N-Methyltransferase in Obesity and Type 2 Diabetes. BioMed Res. Int. 2021, 2021, 9924314. [Google Scholar] [CrossRef]
- Kannt, A.; Pfenninger, A.; Teichert, L.; Tönjes, A.; Dietrich, A.; Schön, M.R.; Klöting, N.; Blüher, M. Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance. Diabetologia 2015, 58, 799–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, R.B.; Facey, P.D. Nicotinamide N-Methyltransferase: An Emerging Protagonist in Cancer Macro(r)evolution. Biomolecules 2021, 11, 1418. [Google Scholar] [CrossRef]
- Ogawa, M.; Tanaka, A.; Namba, K.; Shia, J.; Wang, J.Y.; Roehrl, M.H.A. Tumor stromal nicotinamide N-methyltransferase overexpression as a prognostic biomarker for poor clinical outcome in early-stage colorectal cancer. Sci. Rep. 2022, 12, 2767. [Google Scholar] [CrossRef]
- Sun, W.; Zou, Y.; Cai, Z.; Huang, J.; Hong, X.; Liang, Q.; Jin, W. Overexpression of NNMT in Glioma Aggravates Tumor Cell Progression: An Emerging Therapeutic Target. Cancers 2022, 14, 3538. [Google Scholar] [CrossRef]
- Kocinaj, A.; Chaudhury, T.; Uddin, M.S.; Junaid, R.R.; Ramsden, D.B.; Hondhamuni, G.; Klamt, F.; Parsons, L.; Parsons, R.B. High Expression of Nicotinamide N-Methyltransferase in Patients with Sporadic Alzheimer’s Disease. Mol. Neurobiol. 2021, 58, 1769–1781. [Google Scholar] [CrossRef]
- Parsons, R.B.; Smith, S.W.; Waring, R.H.; Williams, A.C.; Ramsden, D.B. High expression of nicotinamide N-methyltransferase in patients with idiopathic Parkinson’s disease. Neurosci. Lett. 2003, 342, 13–16. [Google Scholar] [CrossRef]
- Schmeisser, K.; Parker, J.A. Nicotinamide-N-methyltransferase controls behavior, neurodegeneration and lifespan by regulating neuronal autophagy. PLoS Genet. 2018, 14, e1007561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fedorowicz, A.; Mateuszuk, L.; Kopec, G.; Skórka, T.; Kutryb-Zając, B.; Zakrzewska, A.; Walczak, M.; Jakubowski, A.; Łomnicka, M.; Słomińska, E.; et al. Activation of the nicotinamide N-methyltransferase (NNMT)-1-methylnicotinamide (MNA) pathway in pulmonary hypertension. Respir. Res. 2016, 17, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Q.; Liu, F.; Yang, L.; Fang, Z.; He, J.; Wang, W.; You, P. Lower serum nicotinamide N-methyltransferase levels in patients with bipolar disorder during acute episodes compared to healthy controls: A cross-sectional study. BMC Psychiatry 2020, 20, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bromberg, A.; Lerer, E.; Udawela, M.; Scarr, E.; Dean, B.; Belmaker, R.H.; Ebstein, R.; Agam, G. Nicotinamide-N-methyltransferase (NNMT) in schizophrenia: Genetic association and decreased frontal cortex mRNA levels. Int. J. Neuropsychopharmacol. 2012, 15, 727–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizuno, R.; Hojo, H.; Takahashi, M.; Kashio, S.; Enya, S.; Nakao, M.; Konishi, R.; Yoda, M.; Harata, A.; Hamanishi, J.; et al. Remote solid cancers rewire hepatic nitrogen metabolism via host nicotinamide-N-methyltransferase. Nat. Commun. 2022, 13, 3346. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
DeBalsi, K.L.; Newman, J.H.; Sommerville, L.J.; Phillips, J.A., III; Hamid, R.; Cogan, J.; Fessel, J.P.; Evans, A.M.; Undiagnosed Diseases Network, Undiagnosed Diseases Network; Kennedy, A.D. A Case Study of Dysfunctional Nicotinamide Metabolism in a 20-Year-Old Male. Metabolites 2023, 13, 399. https://doi.org/10.3390/metabo13030399
DeBalsi KL, Newman JH, Sommerville LJ, Phillips JA III, Hamid R, Cogan J, Fessel JP, Evans AM, Undiagnosed Diseases Network Undiagnosed Diseases Network, Kennedy AD. A Case Study of Dysfunctional Nicotinamide Metabolism in a 20-Year-Old Male. Metabolites. 2023; 13(3):399. https://doi.org/10.3390/metabo13030399
Chicago/Turabian StyleDeBalsi, Karen L., John H. Newman, Laura J. Sommerville, John A. Phillips, III, Rizwan Hamid, Joy Cogan, Joshua P. Fessel, Anne M. Evans, Undiagnosed Diseases Network Undiagnosed Diseases Network, and Adam D. Kennedy. 2023. "A Case Study of Dysfunctional Nicotinamide Metabolism in a 20-Year-Old Male" Metabolites 13, no. 3: 399. https://doi.org/10.3390/metabo13030399
APA StyleDeBalsi, K. L., Newman, J. H., Sommerville, L. J., Phillips, J. A., III, Hamid, R., Cogan, J., Fessel, J. P., Evans, A. M., Undiagnosed Diseases Network, Undiagnosed Diseases Network, & Kennedy, A. D. (2023). A Case Study of Dysfunctional Nicotinamide Metabolism in a 20-Year-Old Male. Metabolites, 13(3), 399. https://doi.org/10.3390/metabo13030399