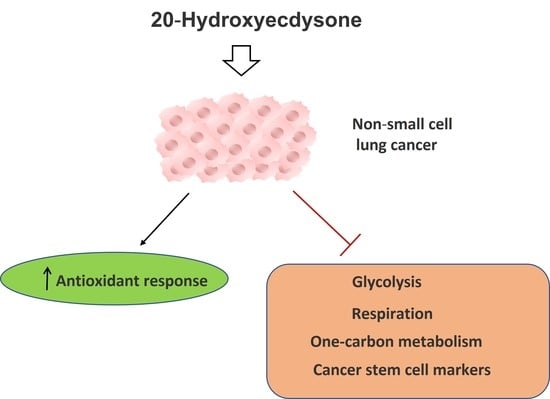
20-Hydroxyecdysone Confers Antioxidant and Antineoplastic Properties in Human Non-Small Cell Lung Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Reagents
2.2. Measurement of ROS Level
2.3. Real-Time PCR
2.4. Qiagen RT2 Profiler™ PCR Array
2.5. RNA-Seq Analysis
2.6. Western-Blot
2.7. SeaHorse Energy Profiling
2.8. ATP Production Assay
2.9. MTT Assay
2.10. Analysis of Drug Synergy
2.11. Bioinformatic Analysis of Lung Cancer Patients’ Survival Rates
2.12. Colony-Formation Assay
2.13. Cell Cycle Analysis
2.14. Annexin V Test
2.15. Study of the Effect of 20E on CSC’s Markers
2.16. Statistical Analysis
3. Results
3.1. 20E Down-Regulates ROS Levels and Induces the Expression of Antioxidative Response Genes
3.2. 20E Slightly Inhibits the Growth of NSCLC Cell Lines
3.3. RNA-Seq Analysis of the 20E-Mediated Impact on Gene Expression
3.4. 20E Down-Regulates Enzymes of Glycolysis and One-Carbon Metabolism
3.5. 20E Inhibits Glycolysis and Respiration in NSCLC Cell Lines
3.6. 20E Sensitizes Lung Cancer Cells to Metabolic Inhibitors
3.7. The Increased Expression of 20E-Suppresed Metabolic Genes Is Associated with the Shortened Survival of Lung Cancer Patients
3.8. 20E Suppressed the Expression of Genes Associated with Cancer Stem-like Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dinan, L.; Dioh, W.; Veillet, S.; Lafont, R. 20-Hydroxyecdysone, from plant extracts to clinical use: Therapeutic potential for the treatment of neuromuscular, cardio-metabolic and respiratory diseases. Biomedicines 2021, 9, 492. [Google Scholar] [CrossRef] [PubMed]
- Cahlíková, L.; Macáková, K.; Chlebek, J.; Hošt’álková, A.; Kulhánková, A.; Opletal, L. Ecdysterone and its activity on some degenerative diseases. Nat. Prod. Commun. 2011, 6, 707–718. [Google Scholar] [CrossRef] [Green Version]
- Latil, M.; Camelo, S.; Veillet, S.; Lafont, R.; Dilda, P.J. Developing new drugs that activate the protective arm of the renin–angiotensin system as a potential treatment for respiratory failure in COVID-19 patients. Drug Discov. Today 2021, 26, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Isenmann, E.; Ambrosio, G.; Joseph, J.F.; Mazzarino, M.; de la Torre, X.; Zimmer, P.; Kazlauskas, R.; Goebel, C.; Botrè, F.; Diel, P. Ecdysteroids as non-conventional anabolic agent: Performance enhancement by ecdysterone supplementation in humans. Arch. Toxicol. 2019, 93, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, S.; Nishimoto, N.; Matsuda, H. Pharmacology of ecdysones in vertebrates. In Invertebrate Endocrinology and Hormonal Heterophylly; Springer: Berlin/Heidelberg, Germany, 1974; pp. 341–344. [Google Scholar]
- Timofeev, N. Leuzea Carthamoides DC.: Application Prospects As. In Functional Foods for Chronic Diseases; D&A Inc.: Richardson, TX, USA, 2006; p. 105. [Google Scholar]
- Hunyadi, A.; Herke, I.; Lengyel, K.; Báthori, M.; Kele, Z.; Simon, A.; Tóth, G.; Szendrei, K. Ecdysteroid-containing food supplements from Cyanotis arachnoidea on the European market: Evidence for spinach product counterfeiting. Sci. Rep. 2016, 6, 37322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Zhao, S.; Ren, L.; Wang, R.; Bai, X.; Han, H.; Li, B.; Chen, H. Research on relationship between tissue quantitative distribution of 3H-Achyranthes bidentata ecdysterone and channel-tropism of herbal drugs in mice. Zhongguo Zhong Yao Za Zhi=Zhongguo Zhongyao Zazhi=China J. Chin. Mater. Med. 2011, 36, 3018–3022. [Google Scholar]
- Dinan, L.; Balducci, C.; Guibout, L.; Foucault, A.-S.; Bakrim, A.; Kumpun, S.; Girault, J.-P.; Tourette, C.; Dioh, W.; Dilda, P. Ecdysteroid metabolism in mammals: The fate of ingested 20-hydroxyecdysone in mice and rats. J. Steroid Biochem. Mol. Biol. 2021, 212, 105896. [Google Scholar] [CrossRef]
- Lafont, R.; Serova, M.; Didry-Barca, B.; Raynal, S.; Guibout, L.; Dinan, L.; Veillet, S.; Latil, M.; Dioh, W.; Dilda, P.J. 20-Hydroxyecdysone activates the protective arm of the RAAS via the MAS receptor. J. Mol. Endocrinol. 2022, 68, 77–87. [Google Scholar] [CrossRef]
- Parr, M.K.; Zhao, P.; Haupt, O.; Ngueu, S.T.; Hengevoss, J.; Fritzemeier, K.H.; Piechotta, M.; Schlörer, N.; Muhn, P.; Zheng, W.Y. Estrogen receptor beta is involved in skeletal muscle hypertrophy induced by the phytoecdysteroid ecdysterone. Mol. Nutr. Food Res. 2014, 58, 1861–1872. [Google Scholar] [CrossRef]
- Shuvalov, O.; Fedorova, O.; Tananykina, E.; Gnennaya, Y.; Daks, A.; Petukhov, A.; Barlev, N. An arthropod hormone, ecdysterone, inhibits the growth of breast cancer cells via different mechanisms. Front. Pharmacol. 2020, 11, 561537. [Google Scholar] [CrossRef]
- Martins, A.; Tóth, N.; Ványolós, A.; Béni, Z.; Zupkó, I.; Molnár, J.; Báthori, M.; Hunyadi, A. Significant activity of ecdysteroids on the resistance to doxorubicin in mammalian cancer cells expressing the human ABCB1 transporter. J. Med. Chem. 2012, 55, 5034–5043. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.; Sipos, P.; Dér, K.; Csábi, J.; Miklos, W.; Berger, W.; Zalatnai, A.; Amaral, L.; Molnár, J.; Szabó-Révész, P. Ecdysteroids sensitize MDR and non-MDR cancer cell lines to doxorubicin, paclitaxel, and vincristine but tend to protect them from cisplatin. BioMed Res. Int. 2015, 2015, 895360. [Google Scholar] [CrossRef] [Green Version]
- Konovalova, N.; Mitrokhin, Y.I.; Volkova, L.; Sidorenko, L.; Todorov, I. Ecdysterone modulates antitumor activity of cytostatics and biosynthesis of macromolecules in tumor-bearing mice. Biol. Bull. Russ. Acad. Sci. 2002, 29, 530–536. [Google Scholar] [CrossRef]
- Romaniuk-Drapała, A.; Lisiak, N.; Totoń, E.; Matysiak, A.; Nawrot, J.; Nowak, G.; Kaczmarek, M.; Rybczyńska, M.; Rubiś, B. Proapoptotic and proautophagic activity of 20-hydroxyecdysone in breast cancer cells in vitro. Chem.-Biol. Interact. 2021, 342, 109479. [Google Scholar] [CrossRef] [PubMed]
- Vágvölgyi, M.; Martins, A.; Kulmány, Á.; Zupkó, I.; Gáti, T.; Simon, A.; Tóth, G.; Hunyadi, A. Nitrogen-containing ecdysteroid derivatives vs. multi-drug resistance in cancer: Preparation and antitumor activity of oximes, oxime ethers and a lactam. Eur. J. Med. Chem. 2018, 144, 730–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bortolozzi, R.; Luraghi, A.; Mattiuzzo, E.; Sacchetti, A.; Silvani, A.; Viola, G. Ecdysteroid derivatives that reverse P-glycoprotein-mediated drug resistance. J. Nat. Prod. 2020, 83, 2434–2446. [Google Scholar] [CrossRef]
- Savchenko, R.G.; Nové, M.; Spengler, G.; Hunyadi, A.; Parfenova, L.V. In vitro adjuvant antitumor activity of various classes of semi-synthetic poststerone derivatives. Bioorganic Chem. 2021, 106, 104485. [Google Scholar] [CrossRef]
- Shuvalov, O.; Daks, A.; Fedorova, O.; Petukhov, A.; Barlev, N. Linking metabolic reprogramming, plasticity and tumor progression. Cancers 2021, 13, 762. [Google Scholar] [CrossRef]
- Mendes, C.; Serpa, J. Metabolic remodelling: An accomplice for new therapeutic strategies to fight lung cancer. Antioxidants 2019, 8, 603. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, M.; Liu, H.; Chen, J. Tumor metabolic reprogramming in lung cancer progression. Oncol. Lett. 2022, 24, 287. [Google Scholar] [CrossRef]
- Vanhove, K.; Graulus, G.-J.; Mesotten, L.; Thomeer, M.; Derveaux, E.; Noben, J.-P.; Guedens, W.; Adriaensens, P. The metabolic landscape of lung cancer: New insights in a disturbed glucose metabolism. Front. Oncol. 2019, 9, 1215. [Google Scholar] [CrossRef] [Green Version]
- Dowling, C.M.; Zhang, H.; Chonghaile, T.N.; Wong, K.-K. Shining a light on metabolic vulnerabilities in non-small cell lung cancer. Biochim. Et Biophys. Acta (BBA) Rev. Cancer 2021, 1875, 188462. [Google Scholar] [CrossRef] [PubMed]
- Shuvalov, O.; Kizenko, A.; Petukhov, A.; Fedorova, O.; Daks, A.; Bottrill, A.; Snezhkina, A.V.; Kudryavtseva, A.V.; Barlev, N. SEMG1/2 augment energy metabolism of tumor cells. Cell Death Dis. 2020, 11, 1047. [Google Scholar] [CrossRef] [PubMed]
- Shuvalov, O.; Kizenko, A.; Shakirova, A.; Fedorova, O.; Petukhov, A.; Aksenov, N.; Vasileva, E.; Daks, A.; Barlev, N. Nutlin sensitizes lung carcinoma cells to interferon-alpha treatment in MDM2-dependent but p53-independent manner. Biochem. Biophys. Res. Commun. 2018, 495, 1233–1239. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-based survival analysis tool tailored for medical research (KMplot): Development and implementation. J. Med. Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef]
- Selby, M.; Delosh, R.; Laudeman, J.; Ogle, C.; Reinhart, R.; Silvers, T.; Lawrence, S.; Kinders, R.; Parchment, R.; Teicher, B.A. 3D models of the NCI60 cell lines for screening oncology compounds. Slas Discov. Adv. Life Sci. RD 2017, 22, 473–483. [Google Scholar] [CrossRef] [Green Version]
- Kirtonia, A.; Sethi, G.; Garg, M. The multifaceted role of reactive oxygen species in tumorigenesis. Cell. Mol. Life Sci. 2020, 77, 4459–4483. [Google Scholar] [CrossRef]
- Kuzmenko, A.I.; Niki, E.; Noguchi, N. New functions of 20-hydroxyecdysone in lipid peroxidation. J. Oleo Sci. 2001, 50, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.-J.; Dai, J.-Q.; Fang, J.-G.; Ma, L.-P.; Hou, L.-F.; Yang, L.; Liu, Z.-L. Antioxidative and free radical scavenging effects of ecdysteroids from Serratula strangulata. Can. J. Physiol. Pharmacol. 2002, 80, 1187–1194. [Google Scholar] [CrossRef]
- Gholipour, P.; Komaki, A.; Ramezani, M.; Parsa, H. Effects of the combination of high-intensity interval training and Ecdysterone on learning and memory abilities, antioxidant enzyme activities, and neuronal population in an Amyloid-beta-induced rat model of Alzheimer’s disease. Physiol. Behav. 2022, 251, 113817. [Google Scholar] [CrossRef]
- Yang, L.; Friedemann, T.; Pan, J. Ecdysterone attenuates the development of radiation-induced oral mucositis in rats at early stage. Radiat. Res. 2021, 196, 366–374. [Google Scholar] [CrossRef] [PubMed]
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The role of Notch3 signaling in cancer stemness and chemoresistance: Molecular mechanisms and targeting strategies. Front. Mol. Biosci. 2021, 8, 694141. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.L.; Gökmen-Polar, Y. HSF1 as a cancer biomarker and therapeutic target. Curr. Cancer Drug Targets 2019, 19, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Silva-Lima, B.; Videira, M. Akt/mTOR Activation in Lung Cancer Tumorigenic Regulators and Their Potential Value as Biomarkers. Onco 2022, 2, 36–55. [Google Scholar] [CrossRef]
- Wang, L.; Hu, F.; Shen, S.; Xiao, H.; Li, G.; Wang, M.; Mei, J. Knockdown of SOX12 expression inhibits the proliferation and metastasis of lung cancer cells. Am. J. Transl. Res. 2017, 9, 4003. [Google Scholar] [PubMed]
- Jiao, X.; Gao, W.; Ren, H.; Wu, Y.; Li, T.; Li, S.; Yan, H. Kruppel like factor 16 promotes lung adenocarcinoma progression by upregulating lamin B2. Bioengineered 2022, 13, 9483–9495. [Google Scholar] [CrossRef] [PubMed]
- Fukushi, A.; Kim, H.-D.; Chang, Y.-C.; Kim, C.-H. Revisited metabolic control and reprogramming cancers by means of the warburg effect in tumor Cells. Int. J. Mol. Sci. 2022, 23, 10037. [Google Scholar] [CrossRef]
- Rosenzweig, A.; Blenis, J.; Gomes, A.P. Beyond the Warburg effect: How do cancer cells regulate one-carbon metabolism? Front. Cell Dev. Biol. 2018, 6, 90. [Google Scholar] [CrossRef]
- Parma, B.; Ramesh, V.; Gollavilli, P.N.; Siddiqui, A.; Pinna, L.; Schwab, A.; Marschall, S.; Zhang, S.; Pilarsky, C.; Napoli, F. Metabolic impairment of non-small cell lung cancers by mitochondrial HSPD1 targeting. J. Exp. Clin. Cancer Res. 2021, 40, 248. [Google Scholar] [CrossRef]
- Yu, T.; Liu, Y.; Xue, J.; Sun, X.; Zhu, D.; Ma, L.; Guo, Y.; Jin, T.; Cao, H.; Chen, Y. Gankyrin modulated non-small cell lung cancer progression via glycolysis metabolism in a YAP1-dependent manner. Cell Death Discov. 2022, 8, 312. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, Y.; Chai, L.; Bu, H.; Yang, Y.; Huang, H.; Ran, J.; Zhu, Y.; Li, L.; Chen, F. FRK plays an oncogenic role in non-small cell lung cancer by enhancing the stemness phenotype via induction of metabolic reprogramming. Int. J. Cancer 2020, 146, 208–222. [Google Scholar] [CrossRef]
- Goetzman, E.S.; Prochownik, E.V. The role for Myc in coordinating glycolysis, oxidative phosphorylation, glutaminolysis, and fatty acid metabolism in normal and neoplastic tissues. Front. Endocrinol. 2018, 9, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cargill, K.R.; Stewart, C.A.; Park, E.M.; Ramkumar, K.; Gay, C.M.; Cardnell, R.J.; Wang, Q.; Diao, L.; Shen, L.; Fan, Y.-H. Targeting MYC-enhanced glycolysis for the treatment of small cell lung cancer. Cancer Metab. 2021, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Fan, M.; Liu, Z.; Li, X.; Wang, H. Serine, glycine and one-carbon metabolism in cancer. Int. J. Oncol. 2021, 58, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Gracilla, D.; Cai, L.; Zhang, M.; Yu, X.; Chen, X.; Zhang, J.; Long, X.; Ding, H.-F.; Yan, C. ATF3 promotes the serine synthesis pathway and tumor growth under dietary serine restriction. Cell Rep. 2021, 36, 109706. [Google Scholar] [CrossRef] [PubMed]
- Tajan, M.; Hennequart, M.; Cheung, E.C.; Zani, F.; Hock, A.K.; Legrave, N.; Maddocks, O.D.; Ridgway, R.A.; Athineos, D.; Suárez-Bonnet, A. Serine synthesis pathway inhibition cooperates with dietary serine and glycine limitation for cancer therapy. Nat. Commun. 2021, 12, 366. [Google Scholar] [CrossRef]
- Sorge, S.; Theelke, J.; Yildirim, K.; Hertenstein, H.; McMullen, E.; Müller, S.; Altbürger, C.; Schirmeier, S.; Lohmann, I. ATF4-induced warburg metabolism drives over-proliferation in Drosophila. Cell Rep. 2020, 31, 107659. [Google Scholar] [CrossRef]
- Chou, T.; Martin, N. CompuSyn for Drug Combinations: PC Software and User’s Guide: A Computer Program for Quantitation of Synergism and Antagonism in Drug Combinations, and the Determination of IC50 and ED50 and LD50 Values; ComboSyn: Paramus, NJ, USA, 2005. [Google Scholar]
- Zheng, Y.; Wang, L.; Yin, L.; Yao, Z.; Tong, R.; Xue, J.; Lu, Y. Lung cancer stem cell markers as therapeutic targets: An update on signaling pathways and therapies. Front. Oncol. 2022, 12, 873994. [Google Scholar] [CrossRef]
- Prabavathy, D.; Swarnalatha, Y.; Ramadoss, N. Lung cancer stem cells—Origin, characteristics and therapy. Stem Cell Investig. 2018, 5, 6. [Google Scholar] [CrossRef] [Green Version]
- Levina, V.; Marrangoni, A.; Wang, T.; Parikh, S.; Su, Y.; Herberman, R.; Lokshin, A.; Gorelik, E. Elimination of human lung cancer stem cells through targeting of the stem cell factor–c-kit autocrine signaling loop. Cancer Res. 2010, 70, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Jaramillo-Rangel, G.; Chávez-Briones, M.-d.-L.; Ancer-Arellano, A.; Ortega-Martínez, M. Nestin-expressing cells in the lung: The bad and the good parts. Cells 2021, 10, 3413. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef] [PubMed]
- Zaidieh, T.; Smith, J.R.; Ball, K.E.; An, Q. ROS as a novel indicator to predict anticancer drug efficacy. BMC Cancer 2019, 19, 1224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, H.; Takada, K. Reactive oxygen species in cancer: Current findings and future directions. Cancer Sci. 2021, 112, 3945–3952. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, D.; Huang, L.; Wang, S.; Jin, Y. Targeting reactive oxygen species capacity of tumor cells with repurposed drug as an anticancer therapy. Oxidative Med. Cell. Longev. 2021, 2021, 8532940. [Google Scholar] [CrossRef]
- Biswas, P.; Dey, D.; Biswas, P.K.; Rahaman, T.I.; Saha, S.; Parvez, A.; Khan, D.A.; Lily, N.J.; Saha, K.; Sohel, M. A comprehensive analysis and anti-cancer activities of quercetin in ROS-mediated cancer and cancer stem cells. Int. J. Mol. Sci. 2022, 23, 11746. [Google Scholar] [CrossRef]
- Imran, M.; Salehi, B.; Sharifi-Rad, J.; Aslam Gondal, T.; Saeed, F.; Imran, A.; Shahbaz, M.; Tsouh Fokou, P.V.; Umair Arshad, M.; Khan, H. Kaempferol: A key emphasis to its anticancer potential. Molecules 2019, 24, 2277. [Google Scholar] [CrossRef] [Green Version]
- Satari, A.; Ghasemi, S.; Habtemariam, S.; Asgharian, S.; Lorigooini, Z. Rutin: A flavonoid as an effective sensitizer for anticancer therapy; insights into multifaceted mechanisms and applicability for combination therapy. Evid.-Based Complement. Altern. Med. 2021, 2021, 9913179. [Google Scholar] [CrossRef]
- Quarta, A.; Gaballo, A.; Pradhan, B.; Patra, S.; Jena, M.; Ragusa, A. Beneficial oxidative stress-related trans-resveratrol effects in the treatment and prevention of breast cancer. Appl. Sci. 2021, 11, 11041. [Google Scholar] [CrossRef]
- Slika, H.; Mansour, H.; Wehbe, N.; Nasser, S.A.; Iratni, R.; Nasrallah, G.; Shaito, A.; Ghaddar, T.; Kobeissy, F.; Eid, A.H. Therapeutic potential of flavonoids in cancer: ROS-mediated mechanisms. Biomed. Pharmacother. 2022, 146, 112442. [Google Scholar] [CrossRef]
- Ganapathy-Kanniappan, S. Molecular intricacies of aerobic glycolysis in cancer: Current insights into the classic metabolic phenotype. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 667–682. [Google Scholar] [CrossRef] [PubMed]
- Shuvalov, O.; Petukhov, A.; Daks, A.; Fedorova, O.; Vasileva, E.; Barlev, N.A. One-carbon metabolism and nucleotide biosynthesis as attractive targets for anticancer therapy. Oncotarget 2017, 8, 23955–23977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, P.; Yang, Q. Overexpression of SHMT2 predicts a poor prognosis and promotes tumor cell growth in bladder cancer. Front. Genet. 2021, 12, 682856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, S.; Zhou, H.; Li, R.; Xia, X.; Xiong, H. Identification of MTHFD2 as a prognostic biomarker and ferroptosis regulator in triple-negative breast cancer. Front. Oncol. 2023, 13, 1098357. [Google Scholar] [CrossRef]
- Zhu, J.; Ma, J.; Wang, X.; Ma, T.; Zhang, S.; Wang, W.; Zhou, X.; Shi, J. High Expression of PHGDH Predicts Poor Prognosis in Non–Small Cell Lung Cancer. Transl. Oncol. 2016, 9, 592–599. [Google Scholar] [CrossRef]
- Huang, M.-Y.; Liu, X.-Y.; Shao, Q.; Zhang, X.; Miao, L.; Wu, X.-Y.; Xu, Y.-X.; Wang, F.; Wang, H.-Y.; Zeng, L. Phosphoserine phosphatase as a prognostic biomarker in patients with gastric cancer and its potential association with immune cells. BMC Gastroenterol. 2022, 22, 1. [Google Scholar] [CrossRef]
- Mingtao, F.; Huanhuan, C.; Wenjing, T.; Liangdong, L.; Yang, G.; Lei, C.; Deheng, L.; Xin, C.; Fengfeng, X.; Changshuai, Z. An integrated pan-cancer analysis of PSAT1: A potential biomarker for survival and immunotherapy. Front. Genet. 2022, 13, 975381. [Google Scholar]
- Wang, M.; Lu, Y.; Wang, H.; Wu, Y.; Xu, X.; Li, Y. High ATF4 expression is associated with poor prognosis, amino acid metabolism, and autophagy in gastric cancer. Front. Oncol. 2021, 11, 740120. [Google Scholar] [CrossRef]
- He, W.-L.; Weng, X.-T.; Wang, J.-L.; Lin, Y.-K.; Liu, T.-W.; Zhou, Q.-Y.; Hu, Y.; Pan, Y.; Chen, X.-L. Association between c-Myc and colorectal cancer prognosis: A meta-analysis. Front. Physiol. 2018, 9, 1549. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Mei, S.; Ding, Q.; Wang, Q.; Yu, L.; Zi, F. A pan-cancer analysis of the role of hexokinase II (HK2) in human tumors. Sci. Rep. 2022, 12, 18807. [Google Scholar] [CrossRef]
- Yu, C.; Hou, L.; Cui, H.; Zhang, L.; Tan, X.; Leng, X.; Li, Y. LDHA upregulation independently predicts poor survival in lung adenocarcinoma, but not in lung squamous cell carcinoma. Future Oncol. 2018, 14, 2483–2492. [Google Scholar] [CrossRef] [PubMed]
- Marbaniang, C.; Kma, L. Dysregulation of glucose metabolism by oncogenes and tumor suppressors in cancer cells. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 2377. [Google Scholar] [PubMed]
- Daks, A.; Shuvalov, O.; Fedorova, O.; Petukhov, A.; Lezina, L.; Zharova, A.; Baidyuk, E.; Khudiakov, A.; Barlev, N.A. p53-independent effects of Set7/9 lysine methyltransferase on metabolism of non-small cell lung cancer cells. Front. Oncol. 2021, 11, 3985. [Google Scholar] [CrossRef]
- Sheng, H.; Tang, W. Glycolysis inhibitors for anticancer therapy: A review of recent patents. Recent Pat. Anti-Cancer Drug Discov. 2016, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sun, G.; Sun, X.; Li, F.; Zhao, L.; Zhong, R.; Peng, Y. The potential of lonidamine in combination with chemotherapy and physical therapy in cancer treatment. Cancers 2020, 12, 3332. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Sun, G.; Sun, X.; Zhao, L.; Zhong, R.; Peng, Y. Tumor energy metabolism and potential of 3-bromopyruvate as an inhibitor of aerobic glycolysis: Implications in tumor treatment. Cancers 2019, 11, 317. [Google Scholar] [CrossRef] [Green Version]
- Cuthbertson, C.R.; Arabzada, Z.; Bankhead III, A.; Kyani, A.; Neamati, N. A review of small-molecule inhibitors of one-carbon enzymes: SHMT2 and MTHFD2 in the spotlight. ACS Pharmacol. Transl. Sci. 2021, 4, 624–646. [Google Scholar] [CrossRef]
- Liu, C.; Jin, Y.; Fan, Z. The mechanism of Warburg effect-induced chemoresistance in cancer. Front. Oncol. 2021, 11, 698023. [Google Scholar] [CrossRef]
- Walcher, L.; Kistenmacher, A.-K.; Suo, H.; Kitte, R.; Dluczek, S.; Strauß, A.; Blaudszun, A.-R.; Yevsa, T.; Fricke, S.; Kossatz-Boehlert, U. Cancer stem cells—Origins and biomarkers: Perspectives for targeted personalized therapies. Front. Immunol. 2020, 11, 1280. [Google Scholar] [CrossRef]
- Chang, J.C. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine 2016, 95 (Suppl. S1), S20–S25. [Google Scholar] [CrossRef]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Han, Z.; Zhu, Y.; Chen, J.; Li, W. The role and specific mechanism of OCT4 in cancer stem cells: A review. Int. J. Stem Cells 2020, 13, 312–325. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Ma, Y.; Yang, Y.; Zhang, L.; Han, H.; Chen, J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol. Lett. 2018, 15, 5627–5633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codony-Servat, J.; Codony-Servat, C.; Cardona, A.F.; Giménez-Capitán, A.; Drozdowskyj, A.; Berenguer, J.; Bracht, J.W.P.; Ito, M.; Karachaliou, N.; Rosell, R. Cancer stem cell biomarkers in egfr-mutation–positive non–small-cell lung cancer. Clin. Lung Cancer 2019, 20, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Kurtti, T.J.; Munderloh, U.G. The effects of 20-hydroxyecdysone and juvenile hormone III on tick cells. J. Parasitol. 1983, 69, 1072–1078. [Google Scholar] [CrossRef]
- Lafont, R.; Raynal, S.; Serova, M.; Didry-Barca, B.; Guibout, L.; Latil, M.; Dilda, P.J.; Dioh, W.; Veillet, S. 20-Hydroxyecdysone activates the protective arm of the renin angiotensin system via Mas receptor. BioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Bounias, M. Biphasic regulation in ligand-receptor interactions. J. Enzym. Inhib. 1990, 3, 323–326. [Google Scholar] [CrossRef]
- Calabrese, E.J. Biphasic dose responses in biology, toxicology and medicine: Accounting for their generalizability and quantitative features. Environ. Pollut. 2013, 182, 452–460. [Google Scholar] [CrossRef]











| Dose 20E | Dose 2nd Compound | A549 | H460 | ||
|---|---|---|---|---|---|
| Effect | CI | Effect | CI | ||
| 0.1 µM | 2-DG (2.5 mM) | 0.64 | 0.53 | 0.62 | 0.68 |
| 1 µM | 2-DG (2.5 mM) | 0.57 | 0.42 | 0.56 | 0.59 |
| 10 µM | 2-DG (2.5 mM) | 0.51 | 0.37 | 0.49 | 0.51 |
| 0.1 µM | MF (5 mM) | 0.72 | 0.85 | 0.82 | 0.59 |
| 1 µM | MF (5 mM) | 0.61 | 0.52 | 0.81 | 0.57 |
| 10 µM | MF (5 mM) | 0.55 | 0.44 | 0.79 | 0.52 |
| 0.1 µM | 3-BP (100 µM) | 0.69 | 0.71 | 0.71 | 0.69 |
| 1 µM | 3-BP (100 µM) | 0.63 | 0.63 | 0.65 | 0.59 |
| 10 µM | 3-BP (100 µM) | 0.58 | 0.61 | 0.56 | 0.48 |
| 0.1 µM | Gemc (500 nM) | 0.67 | 0.91 | 0.75 | 0.6 |
| 1 µM | Gemc (500 nM) | 0.57 | 0.59 | 0.75 | 0.6 |
| 10 µM | Gemc (500 nM) | 0.51 | 0.48 | 0.64 | 0.37 |
| 0.1 µM | 5-FU (5 µM) | 0.7 | 0.75 | 0.69 | 0.76 |
| 1 µM | 5-FU (5 µM) | 0.6 | 0.56 | 0.64 | 0.68 |
| 10 µM | 5-FU (5 µM) | 0.58 | 0.57 | 0.64 | 0.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shuvalov, O.; Kirdeeva, Y.; Fefilova, E.; Netsvetay, S.; Zorin, M.; Vlasova, Y.; Fedorova, O.; Daks, A.; Parfenyev, S.; Barlev, N. 20-Hydroxyecdysone Confers Antioxidant and Antineoplastic Properties in Human Non-Small Cell Lung Cancer Cells. Metabolites 2023, 13, 656. https://doi.org/10.3390/metabo13050656
Shuvalov O, Kirdeeva Y, Fefilova E, Netsvetay S, Zorin M, Vlasova Y, Fedorova O, Daks A, Parfenyev S, Barlev N. 20-Hydroxyecdysone Confers Antioxidant and Antineoplastic Properties in Human Non-Small Cell Lung Cancer Cells. Metabolites. 2023; 13(5):656. https://doi.org/10.3390/metabo13050656
Chicago/Turabian StyleShuvalov, Oleg, Yulia Kirdeeva, Elizaveta Fefilova, Sofia Netsvetay, Mark Zorin, Yulia Vlasova, Olga Fedorova, Alexandra Daks, Sergey Parfenyev, and Nickolai Barlev. 2023. "20-Hydroxyecdysone Confers Antioxidant and Antineoplastic Properties in Human Non-Small Cell Lung Cancer Cells" Metabolites 13, no. 5: 656. https://doi.org/10.3390/metabo13050656
APA StyleShuvalov, O., Kirdeeva, Y., Fefilova, E., Netsvetay, S., Zorin, M., Vlasova, Y., Fedorova, O., Daks, A., Parfenyev, S., & Barlev, N. (2023). 20-Hydroxyecdysone Confers Antioxidant and Antineoplastic Properties in Human Non-Small Cell Lung Cancer Cells. Metabolites, 13(5), 656. https://doi.org/10.3390/metabo13050656