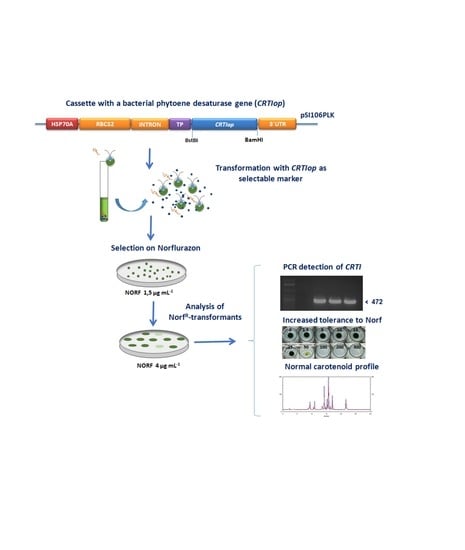
The Bacterial Phytoene Desaturase-Encoding Gene (CRTI) is an Efficient Selectable Marker for the Genetic Transformation of Eukaryotic Microalgae
Abstract
:1. Introduction
2. Results and Discussion
2.1. Construction of Plasmid pSI06PLK-CRTIop
2.2. Sensitivity of the Chlorophyte Microalgae to the Bleaching Herbicide Norflurazon
2.3. Transformation of Chlamydomonas reinhardtii with the Plasmid pSI106PLK-tpCRTIop and Selection of Norflurazon-Resistant NorfR-Chlamydomonas Transformants
2.4. Carotenoid Composition of Norflurazon-Resistant NorfR-Chlamydomonas Transformants
3. Materials and Methods
3.1. Strains and Culture Conditions
3.2. Microalgal Expression of pSI106PLK Plasmid
3.3. Chlamydomonas Nuclear Transformation
3.4. Determination of Carotenoids
3.5. Dry Weight Determination
3.6. Herbicide Sensitivity Test
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Varela, J.C.; Pereira, H.; Vila, M.; León, R. Production of carotenoids by microalgae: Achievements and challenges. Photosynth. Res. 2015, 125, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Valverde, F.; Romero-Campero, F.J.; León, R.; Guerrero, M.G.; Serrano, A. New challenges in microalgae biotechnology. Eur. J. Protistol. 2016, 55, 95–101. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.I.; Shin, J.H.; Kim, J.D. The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factor. 2018, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Schulze, P.S.C.; Guerra, R.; Pereira, H.; Schüler, L.M.; Varela, J.C.S. Flashing LEDs for Microalgal Production. Trends Biotechnol. 2017, 35, 1088–1101. [Google Scholar] [CrossRef] [PubMed]
- Mallick, N.; Bagchi, S.K.; Koley, S.; Singh, A.K. Progress and Challenges in Microalgal Biodiesel Production. Front. Microbiol. 2016, 7, 1019. [Google Scholar] [CrossRef]
- Rengel, R.; Smith, R.T.; Haslam, R.P.; Sayanova, O.; Vila, M.; León, R. Overexpression of acetyl-CoA synthetase (ACS) enhances the biosynthesis of neutral lipids and starch in the green microalga Chlamydomonas reinhardtii. Algal Res. 2018, 31, 183–193. [Google Scholar] [CrossRef]
- Bellou, S.; Triantaphyllidou, I.-E.; Aggeli, D.; Elazzazy, A.M.; Baeshen, M.N.; Aggelis, G. Microbial oils as food additives: Recent approaches for improving microbial oil production and its polyunsaturated fatty acid content. Curr. Opin. Biotechnol. 2016, 37, 24–35. [Google Scholar] [CrossRef]
- Gimpel, J.A.; Henríquez, V.; Mayfield, S.P. In Metabolic Engineering of Eukaryotic Microalgae: Potential and Challenges Come with Great Diversity. Front. Microbiol. 2015, 6, 1376. [Google Scholar] [CrossRef]
- Scranton, M.A.; Ostrand, J.T.; Fields, F.J.; Mayfield, S.P. Chlamydomonas as a model for biofuels and bio-products production. Plant J. 2015, 82, 523–531. [Google Scholar] [CrossRef]
- León, R.; Fernández, E. Nuclear Transformation of Eukaryotic Microalgae. In Transgenic Microalgae as Green Cell Factories; Springer: New York, NY, USA, 2007; Volume 616, pp. 1–11. [Google Scholar]
- Nielsen, K.M.; Bøhn, T.; Townsend, J.P. Detecting rare gene transfer events in bacterial populations. Front. Microbiol. 2014, 4, 415. [Google Scholar] [CrossRef]
- Brueggeman, A.J.; Kuehler, D.; Weeks, D.P. Evaluation of three herbicide resistance genes for use in genetic transformations and for potential crop protection in algae production. Plant Biotechnol. J. 2014, 12, 894–902. [Google Scholar] [CrossRef] [Green Version]
- Kovar, J.L.; Zhang, J.; Funke, R.P.; Weeks, D.P. Molecular analysis of the acetolactate synthase gene of Chlamydomonas reinhardtii and development of a genetically engineered gene as a dominant selectable marker for genetic transformation. Plant J. 2002, 29, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Lapidot, M.; Raveh, D.; Sivan, A.; Arad, S.M.; Shapira, M. Stable chloroplast transformation of the unicellular red alga Porphyridium species. Plant Physiol. 2002, 129, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Grundman, O.; Khozin-Goldberg, I.; Raveh, D.; Cohen, Z.; Vyazmensky, M.; Boussiba, S.; Shapira, M. Cloning, mutagenesis, and characterization of the microalga Parietochloris incisa acetohydroxyacid synthase, and its possible use as an endogenous selection marker. Biotechnol. Bioeng. 2012, 109, 2340–2348. [Google Scholar] [CrossRef]
- Michel, A.; Arias, R.S.; Scheffler, B.E.; Duke, S.O.; Netherland, M.; Dayan, F.E. Somatic mutation-mediated evolution of herbicide resistance in the nonindigenous invasive plant hydrilla (Hydrilla verticillata). Mol. Ecol. 2004, 13, 3229–3237. [Google Scholar] [CrossRef] [PubMed]
- Arias, R.S.; Dayan, F.E.; Michel, A.; Howell, J.; Scheffler, B.E. Characterization of a higher plant herbicide-resistant phytoene desaturase and its use as a selectable marker. Plant Biotechnol. J. 2006, 4, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Breitenbach, J.; Zhu, C.; Sandmann, G. Bleaching Herbicide Norflurazon Inhibits Phytoene Desaturase by Competition with the Cofactors. J. Agric. Food Chem. 2001, 49, 5270–5272. [Google Scholar] [CrossRef] [PubMed]
- Chamovitz, D.; Sandmann, G.; Hirschberg, J. Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 1993, 268, 17348–17353. [Google Scholar]
- Cunningham, F.X.; Gantt, E. Genes And Enzymes of Carotenoid Biosynthesis in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 557–583. [Google Scholar] [CrossRef]
- Sandmann, G.; Römer, S.; Fraser, P.D. Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab. Eng. 2006, 8, 291–302. [Google Scholar] [CrossRef]
- Misawa, N.; Nakagawa, M.; Kobayashi, K.; Yamano, S.; Izawa, Y.; Nakamura, K.; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990, 172, 6704–6712. [Google Scholar] [CrossRef] [PubMed]
- Misawa, N.; Yamano, S.; Linden, H.; de Felipe, M.R.; Lucas, M.; Ikenaga, H.; Sandmann, G. Functional expression of the Erwinia uredovora carotenoid biosynthesis gene crtl in transgenic plants showing an increase of beta-carotene biosynthesis activity and resistance to the bleaching herbicide norflurazon. Plant J. 1993, 4, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Romer, S.; Shipton, C.A.; Mills, P.B.; Kiano, J.W.; Misawa, N.; Drake, R.G.; Schuch, W.; Bramley, P.M. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. USA 2002, 99, 1092–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ducreux, L.J.M.; Morris, W.L.; Hedley, P.E.; Shepherd, T.; Davies, H.V.; Millam, S.; Taylor, M.A. Metabolic engineering of high carotenoid potato tubers containing enhanced levels of beta-carotene and lutein. J. Exp. Bot. 2005, 56, 81–89. [Google Scholar]
- Díaz-Santos, E.; Vila, M.; Vigara, J.; León, R. A new approach to express transgenes in microalgae and its use to increase the flocculation ability of Chlamydomonas reinhardtii. J. Appl. Phycol. 2016, 28, 1611–1621. [Google Scholar] [CrossRef]
- Suarez, J.V.; Banks, S.; Thomas, P.G.; Day, A. A new F131V mutation in Chlamydomonas phytoene desaturase locates a cluster of norflurazon resistance mutations near the FAD-binding site in 3D protein models. PLoS ONE 2014, 9, e99894. [Google Scholar] [CrossRef] [PubMed]
- Steinbrenner, J.; Sandmann, G. Transformation of the green alga Haematococcus pluvialis with a phytoene desaturase for accelerated astaxanthin biosynthesis. Appl. Environ. Microbiol. 2006, 72, 7477–7484. [Google Scholar] [CrossRef]
- Sharon-Gojman, R.; Maimon, E.; Leu, S.; Zarka, A.; Boussiba, S. Advanced methods for genetic engineering of Haematococcus pluvialis (Chlorophyceae, Volvocales). Algal Res. 2015, 10, 8–15. [Google Scholar] [CrossRef]
- Huang, J.; Liu, J.; Li, Y.; Chen, F. Isolation and Characterization of the Phytoene Desaturase Gene As a Potential Selective Marker for Genetic Engineering of the Astaxanthin-Producing Green Alga Chlorella zofingiensis (Chlorophyta). J. Phycol. 2008, 44, 684–690. [Google Scholar] [CrossRef]
- Xue, J.; Niu, Y.-F.; Huang, T.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 2015, 27, 1–9. [Google Scholar] [CrossRef]
- Prasad, B.; Vadakedath, N.; Jeong, H.-J.; General, T.; Cho, M.-G.; Lein, W. Agrobacterium tumefaciens-mediated genetic transformation of haptophytes (Isochrysis species). Appl. Microbiol. Biotechnol. 2014, 98, 8629–8639. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.T.; Sharifi, M.N.; Poddar, S.; Dent, R.M.; Niyogi, K.K. Intragenic Enhancers and Suppressors of Phytoene Desaturase Mutations in Chlamydomonas reinhardtii. PLoS ONE 2012, 7, e42196. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, Z.; Gerken, H.; Huang, J.; Jiang, Y.; Chen, F. Genetic engineering of the green alga Chlorella zofingiensis: A modified norflurazon-resistant phytoene desaturase gene as a dominant selectable marker. Appl. Microbiol. Biotechnol. 2014, 98, 5069–5079. [Google Scholar] [CrossRef] [PubMed]
- Loppes, R.; Radoux, M.; Ohresser, M.C.; Matagne, R.F. Transcriptional regulation of the Nia1 gene encoding nitrate reductase in Chlamydomonas reinhardtii: Effects of various environmental factors on the expression of a reporter gene under the control of the Nia1 promoter. Plant Mol. Biol. 1999, 41, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Guillard, R.R.; Ryther, J.H. Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (cleve) Gran. Can. J. Microbiol. 1962, 8, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.K.; Johnson, E.J.; MacElroy, R.D.; Speer, H.L.; Bruff, B.S. Effects of salts on the halophilic alga Dunaliella viridis. J. Bacteriol. 1968, 95, 1461–1468. [Google Scholar] [PubMed]
- León, R.; Couso, I.; Fernández, E. Metabolic engineering of ketocarotenoids biosynthesis in the unicelullar microalga Chlamydomonas reinhardtii. J. Biotechnol. 2007, 130, 143–152. [Google Scholar] [CrossRef]
- Sizova, I.; Fuhrmann, M.; Hegemann, P. A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 2001, 277, 221–229. [Google Scholar] [CrossRef]
- Kindle, K.L. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 1990, 87, 1228–1232. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]




| Strain | C1 | C2 | T1 | T11 | T19 | T21 |
|---|---|---|---|---|---|---|
| Norflurazon (µg mL−1) | - | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 |
| Phytoene (µg mL−1) | 0.1 ± 0.5 | 4.4 ± 0.4 | 1.8 ± 0.1 | 2.1 ± 0.2 | 2 ± 0.2 | 1.5 ± 0.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molina-Márquez, A.; Vila, M.; Vigara, J.; Borrero, A.; León, R. The Bacterial Phytoene Desaturase-Encoding Gene (CRTI) is an Efficient Selectable Marker for the Genetic Transformation of Eukaryotic Microalgae. Metabolites 2019, 9, 49. https://doi.org/10.3390/metabo9030049
Molina-Márquez A, Vila M, Vigara J, Borrero A, León R. The Bacterial Phytoene Desaturase-Encoding Gene (CRTI) is an Efficient Selectable Marker for the Genetic Transformation of Eukaryotic Microalgae. Metabolites. 2019; 9(3):49. https://doi.org/10.3390/metabo9030049
Chicago/Turabian StyleMolina-Márquez, Ana, Marta Vila, Javier Vigara, Ana Borrero, and Rosa León. 2019. "The Bacterial Phytoene Desaturase-Encoding Gene (CRTI) is an Efficient Selectable Marker for the Genetic Transformation of Eukaryotic Microalgae" Metabolites 9, no. 3: 49. https://doi.org/10.3390/metabo9030049
APA StyleMolina-Márquez, A., Vila, M., Vigara, J., Borrero, A., & León, R. (2019). The Bacterial Phytoene Desaturase-Encoding Gene (CRTI) is an Efficient Selectable Marker for the Genetic Transformation of Eukaryotic Microalgae. Metabolites, 9(3), 49. https://doi.org/10.3390/metabo9030049