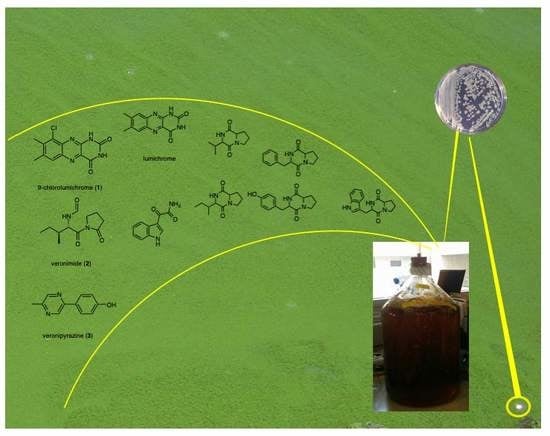
Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom
Abstract
:1. Introduction
2. Results
2.1. Structure Elucidation of 9-Chlorolumichrome (1)
2.2. Structure Elucidation of Veronimide (2)
2.3. Structure Elucidation of Veronipyrazine (3)
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Isolation and Identification of A. veronii Strain A134
4.3. Isolation and Identification of the Metabolites
4.4. Synthesis of 9-Chlorolumichrome
4.5. Determination of the Absolute Configuration of the Amino Acids
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Navarro, A.; Martinez-Murcia, A. Phylogenetic analyses of the genus Aeromonas based on housekeeping gene sequencing and its influence on systematics. J. Appl. Microbiol. 2018, 125, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.D.; Mitchell, R. Antagonism of bacterial extracellular metabolites to freshwater-fouling invertebrate zebra mussels, Dreissena polymorpha. J. Microbiol. 2001, 39, 133–138. [Google Scholar]
- Kamal, S.A.; Hamza, L.F.; Ibraheam, I.A. Characterization of antifungal metabolites produced by Aeromonas hydrophila and analysis of its chemical compounds using GC-MS. Res. J. Pharm. Tech. 2017, 10, 3845–3851. [Google Scholar] [CrossRef]
- Nie, M.; Nie, H.; Cao, W.; Wang, X.; Guo, Y.; Tian, X.; Yin, X.; Wang, Y. Phenanthrene Metabolites from a New Polycyclic Aromatic Hydrocarbon-Degrading Bacterium Aeromonas salmonicida subsp. Achromogenes Strain NY4. Polycyc. Arom. Compd. 2016, 36, 132–151. [Google Scholar] [CrossRef]
- Weiss, G.; Kovalerchick, D.; Murik, O.; De Philippis, R.; Carmeli, S.; Kaplan, A. Increased algicidal activity of Aeromonas veronii in response to Microcystis aeruginosa: Inter-species crosstalk and secondary metabolites synergism. Environ. Microbiol. 2019, 21, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- AntiSMASH Bacterial Version. Available online: https://antismash.secondarymetabolites.org (accessed on 30 December 2018).
- Karrer, P.; Salomon, H.; Schopp, K.; Schlittler, E.; Fritzsche, H. Ein neues Bestralungsproduckt des Lactoflavins: Lumichrom. Helv. Chim. Acta 1934, 17, 1010–1013. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Lumichrome, a larval, metamorphosis-inducing substance in the ascidian Halocynthia roretzi. Eur. J. Biochem. 1999, 264, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Tanemura, K.; Suzuki, T.; Nishida, Y.; Satsumabayashi, K.; Horaguchiy, T. Halogenation of aromatic compounds by N-chloro-, N-bromo- and N-iodosuccinimide. Chem. Lett. 2003, 32, 932–933. [Google Scholar] [CrossRef]
- Marfey, P. Determination of D-amino acids. II. Use of a bifunctional reagent, 1,5-difluoro-2,4-dinitrobenzene. Carlsberg. Res. Commun. 1984, 49, 591–596. [Google Scholar] [CrossRef]
- Kumar, N.; Mohandas, C.; Nambisan, B.; Kumar, D.R.S.; Lankalapalli, R.S. Isolation of proline-based cyclic dipeptidets from Bacillus sp. N strain associated with rhabitid entomopathogenic nematode and its antimicrobial properties. World J. Microbiol. Biotechnol. 2013, 29, 355–364. [Google Scholar] [CrossRef]
- Li, X.; Dobrestov, S.; Xu, Y.; Xiao, X.; Hung, O.S.; Qian, P. Antifouling diketopiperazines produced by a deep-sea bacterium, Streptomyces fungicidicus. Biofouling 2006, 22, 187–194. [Google Scholar] [CrossRef]
- Wardecki, T.; Brotz, E.; De Ford, C.; von Loewenich, F.D.; Rebets, Y.; Tokovenko, B. Endophytic Streptomyces in the traditional medicinal plant Arnica montana L.: Secondary metabolites and biological activity. Antonie van Leeuwenhoek 2015, 108, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Van Pee, K.-H.; Patallo, E.P. Flavin-dependent halogenases involved in secondary metabolism in bacteria. Appl. Microbiol. Biotechnol. 2006, 70, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Schatz, D.; Keren, Y.; Hadas, O.; Carmeli, S.; Sukenik, A.; Kaplan, A. Ecological implications of the emergence of non-toxic subcultures from toxic Microcystis strains. Environm. Microbiol. 2005, 7, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Detwiler, M.M.; Hamp, T.J.; Kazim, A.L. DNA sequencing using the liquid polymer POP-7 ™ on an ABI P. Benchmarks 2004, 36, 932–933. [Google Scholar]


| 9-Chlorolumichrome (1) | Lumichrome | ||||
|---|---|---|---|---|---|
| Position | δC, Mult.b | δH, Mult.c | LRd C-H Correlations | δC, Mult.b | δH, Mult.c |
| 1 | - | 11.74, se | - | - | 11.65, s |
| 2 | 150.2, C | - | - | 150.3, C | - |
| 3 | - | 12.03, s | - | - | 11.82, s |
| 4 | 160.5, C | - | - | 160.8, C | - |
| 4a | 131.5, C | - | 1, 3 | 130.4, C | - |
| 5a | 138.3, C | - | 6 | 138.6, C | - |
| 6 | 127.9, CH | 7.94, s | 11 | 128.9, CH | 7.91, s |
| 7 | 139.0, C | - | 6, 11, 12 | 144.9, C | - |
| 8 | 141.7, C | - | 11, 12 | 139.1, C | - |
| 9 | 128.7, C | - | 6, 12 | 126.0, CH | 7.70, s |
| 9a | 138.2, C | - | 6 | 141.8, C | - |
| 10a | 146.9, C | - | - | 146.7, C | - |
| 11 | 21.0, CH3 | 2.52, s | 6 | 19.8, CH3 | 2.45, s |
| 12 | 17.9, CH3 | 2.57, s | 11 | 20.4, CH3 | 2.48, s |
| Position | δC, Mult.b | δH, Mult.c (J in Hz) | COSY Correlations | LRd C-H Correlations |
|---|---|---|---|---|
| 1 | 175.7, C | - | - | 2a, 2b, 3a, 3b, 4a, 4b |
| 2 | 33.4, CH2 | a 2.61, me b 3.57, me | 3a, 3b | 4a, 4b |
| 3 | 16.9, CH2 | a 1.96, me b 1.92, me | 2a, 2b, 4a, 4b | 2a, 2b, 4a, 4b |
| 4 | 45.7, CH2 | a 3.69, me b 3.65, me | 3a, 3b | 2a, 2b |
| 6 | 172.6, C | - | - | 7 |
| 7 | 54.5, CH | 5.62, ddf (9.1, 4.4) | 8, 12 | 12, 13 |
| 8 | 36.6, CH | 1.79, dddqg (10.0, 4.4, 3.9, 6.8) | 7, 9a, 9b, 11 | 7, 9a, 9b, 10, 11 |
| 9 | 23.2, CH2 | a 1.27, ddqh (13.5, 3.9, 7.4) b 1.08, ddq (13.5, 10.0, 7.4) | 8, 9b, 10 8, 9a, 10 | 7, 9a, 9b, 10, 11 |
| 10 | 11.6, CH3 | 0.79, ti (7.4) | 9a, 9b | 9a, 9b |
| 11 | 16.1, CH3 | 0.85, dj (6.8) | 8 | 7, 9a, 9b |
| 12 | - | 8.26, brdk (9.1) | 7, 13 | - |
| 13 | 161.3, CH | 8.03, sl | 12 | 7 |
| Position | δC, Mult.b (JCH in Hz) | δH, Mult.c (J in Hz) | LRd C-H Correlations |
|---|---|---|---|
| 2 | 143.5, CH (181.4) | 8.50, brse | 11 |
| 3 | 150.8, C | - | 2, 5, 11 |
| 5 | 140.0, CH (185.1) | 8.97, sf | - |
| 6 | 149.0, C | - | 2, 5, 8, 8’ |
| 7 | 127.0, C | - | 9, 9’ |
| 8, 8’ | 128.0, CH × 2 | 7.92, dg × 2 (8.7) | - |
| 9, 9’ | 115.9, CH × 2 | 6.87, d × 2 (8.7) | 9’, 9 |
| 10 | 159.1, C | - | 8, 8’, 9, 9’, 10-OH |
| 10-OH | - | 9.83, brs | - |
| 11 | 20.9, CH3 | 2.48, s | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiss, G.; Kovalerchick, D.; Murik, O.; Sukenik, A.; Kaplan, A.; Carmeli, S. Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom. Metabolites 2019, 9, 110. https://doi.org/10.3390/metabo9060110
Weiss G, Kovalerchick D, Murik O, Sukenik A, Kaplan A, Carmeli S. Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom. Metabolites. 2019; 9(6):110. https://doi.org/10.3390/metabo9060110
Chicago/Turabian StyleWeiss, Gad, Dimitry Kovalerchick, Omer Murik, Assaf Sukenik, Aaron Kaplan, and Shmuel Carmeli. 2019. "Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom" Metabolites 9, no. 6: 110. https://doi.org/10.3390/metabo9060110
APA StyleWeiss, G., Kovalerchick, D., Murik, O., Sukenik, A., Kaplan, A., & Carmeli, S. (2019). Secondary Metabolites of Aeromonas veronii Strain A134 Isolated from a Microcystis aeruginosa Bloom. Metabolites, 9(6), 110. https://doi.org/10.3390/metabo9060110