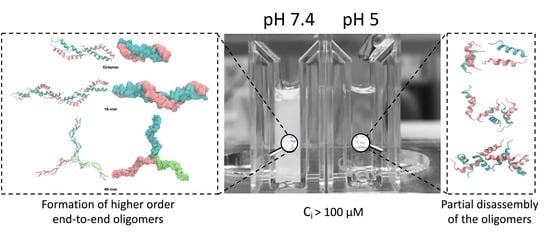
Concentration- and pH-Dependent Oligomerization of the Thrombin-Derived C-Terminal Peptide TCP-25
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptide
2.2. Turbidity Assay
2.3. Electrophoresis and Western Blot
2.4. Circular Dichroism Spectroscopy
2.5. Transmission Electron Microscopy
2.6. Chemical Crosslinking
2.7. High-Pressure Liquid Chromatography (HPLC)
2.8. Thermal and Chemical Denaturation
2.9. Dynamic Light Scattering
2.10. Molecular Modeling and Simulation
2.11. Statistical Analysis
3. Results
3.1. Relationship between Turbidity and Oligomerization/Aggregation of TCP-25
3.2. Structural Changes of TCP-25 Oligomers and Their Organization
3.3. Effects of Oligomerization on Tm and Cm
3.4. Reversibility of Thermal Denaturation of TCP-25
3.5. Size of Oligomers as a Function of Temperature and pH
3.6. Molecular Simulations of TCP-25 Oligomerization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef]
- Sen, C.K.; Gordillo, G.M.; Roy, S.; Kirsner, R.; Lambert, L.; Hunt, T.K.; Gottrup, F.; Gurtner, G.C.; Longaker, M.T. Human skin wounds: A major and snowballing threat to public health and the economy. Wound Repair Regen. 2009, 17, 763–771. [Google Scholar] [CrossRef] [Green Version]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nat. Cell Biol. 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Harder, J.; Gläser, R.; Schröder, J.-M. Review: Human antimicrobial proteins—Effectors of innate immunity. J. Endotoxin Res. 2007, 13, 317–338. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Ganz, T. Cathelicidins: A family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 2002, 9, 18–22. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Genet. 2005, 3, 238–250. [Google Scholar] [CrossRef]
- Easton, D.M.; Nijnik, A.; Mayer, M.L.; Hancock, R.E. Potential of immunomodulatory host defense peptides as novel anti-infectives. Trends Biotechnol. 2009, 27, 582–590. [Google Scholar] [CrossRef]
- Oppenheim, J.J.; Yang, D. Alarmins: Chemotactic activators of immune responses. Curr. Opin. Immunol. 2005, 17, 359–365. [Google Scholar] [CrossRef]
- Pasupuleti, M.; Schmidtchen, A.; Malmsten, M. Antimicrobial peptides: Key components of the innate immune system. Crit. Rev. Biotechnol. 2011, 32, 143–171. [Google Scholar] [CrossRef] [Green Version]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, 11. [Google Scholar] [CrossRef]
- Chen, C.H.; Lu, T.K. Development and Challenges of Antimicrobial Peptides for Therapeutic Applications. Antibiotics 2020, 9, 24. [Google Scholar] [CrossRef] [Green Version]
- Papareddy, P.; Rydengård, V.; Pasupuleti, M.; Walse, B.; Mörgelin, M.; Chalupka, A.; Malmsten, M.; Schmidtchen, A. Proteolysis of Human Thrombin Generates Novel Host Defense Peptides. PLoS Pathog. 2010, 6, e1000857. [Google Scholar] [CrossRef] [Green Version]
- Kalle, M.; Papareddy, P.; Kasetty, G.; Mörgelin, M.; Van Der Plas, M.J.A.; Rydengård, V.; Malmsten, M.; Albiger, B.; Schmidtchen, A. Host Defense Peptides of Thrombin Modulate Inflammation and Coagulation in Endotoxin-Mediated Shock and Pseudomonas aeruginosa Sepsis. PLoS ONE 2012, 7, e51313. [Google Scholar] [CrossRef] [Green Version]
- Kasetty, G.; Papareddy, P.; Kalle, M.; Rydengård, V.; Mörgelin, M.; Albiger, B.; Malmsten, M.; Schmidtchen, A. Structure-Activity Studies and Therapeutic Potential of Host Defense Peptides of Human Thrombin. Antimicrob. Agents Chemother. 2011, 55, 2880–2890. [Google Scholar] [CrossRef] [Green Version]
- Hansen, F.C.; Kalle-Brune, M.; Van Der Plas, M.J.A.; Strömdahl, A.-C.; Malmsten, M.; Mörgelin, M.; Schmidtchen, A. The Thrombin-Derived Host Defense Peptide GKY25 Inhibits Endotoxin-Induced Responses through Interactions with Lipopolysaccharide and Macrophages/Monocytes. J. Immunol. 2015, 194, 5397–5406. [Google Scholar] [CrossRef]
- Saravanan, R.; A Holdbrook, D.; Petrlova, J.; Singh, S.; A Berglund, N.; Choong, Y.K.; Kjellström, S.; Bond, P.J.; Malmsten, M.; Schmidtchen, A. Structural basis for endotoxin neutralisation and anti-inflammatory activity of thrombin-derived C-terminal peptides. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Hansen, F.C.; Strömdahl, A.-C.; Mörgelin, M.; Schmidtchen, A.; Van Der Plas, M.J.A. Thrombin-Derived Host-Defense Peptides Modulate Monocyte/Macrophage Inflammatory Responses to Gram-Negative Bacteria. Front. Immunol. 2017, 8, 843. [Google Scholar] [CrossRef] [Green Version]
- Lim, C.H.; Puthia, M.; Butrym, M.; Tay, H.M.; Lee, M.Z.Y.; Hou, H.W.; Schmidtchen, A. Thrombin-derived host defence peptide modulates neutrophil rolling and migration in vitro and functional response in vivo. Sci. Rep. 2017, 7, 11201. [Google Scholar] [CrossRef] [Green Version]
- Puthia, M.; Butrym, M.; Petrlova, J.; Strömdahl, A.-C.; Andersson, M.Å.; Kjellström, S.; Schmidtchen, A. A dual-action peptide-containing hydrogel targets wound infection and inflammation. Sci. Transl. Med. 2020, 12, eaax6601. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Marchand, C.H.; Malferrari, M.; Murail, S.; Bonacchi, S.; Genovese, D.; Montalti, M.; Venturoli, G.; Falini, G.; Baaden, M.; et al. Glutathionylation primes soluble glyceraldehyde-3-phosphate dehydrogenase for late collapse into insoluble aggregates. Proc. Natl. Acad. Sci. USA 2019, 116, 26057–26065. [Google Scholar] [CrossRef] [PubMed]
- Morrisett, J.D.; David, J.S.K.; Pownall, H.J.; Gotto, A.M. Interaction of an apolipoprotein (apoLP-alanine) with phosphatidylcholine. Biochemistry 1973, 12, 1290–1299. [Google Scholar] [CrossRef] [PubMed]
- Petrlova, J.; Petruk, G.; Huber, R.G.; McBurnie, E.W.; Van Der Plas, M.J.A.; Bond, P.J.; Puthia, M.; Schmidtchen, A. Thrombin-derived C-terminal fragments aggregate and scavenge bacteria and their proinflammatory products. J. Biol. Chem. 2020, 295, 3417–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phadte, A.S.; Santhoshkumar, P.; Sharma, K.K. Characterization of an N-terminal mutant of αA-crystallin αA–R21Q associated with congenital cataract. Exp. Eye Res. 2018, 174, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Eramian, D.; Shen, M.-Y.; Devos, D.; Melo, F.; Sali, A.; Martí-Renom, M.A. A composite score for predicting errors in protein structure models. Protein Sci. 2006, 15, 1653–1666. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, G.; Ramakrishnan, C.; Sasisekharan, V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963, 7, 95–99. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef]
- Huang, J.; MacKerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef]
- Lee, J.; Cheng, X.; Swails, J.M.; Yeom, M.S.; Eastman, P.K.; Lemkul, J.A.; Wei, S.; Buckner, J.; Jeong, J.C.; Qi, Y.; et al. CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Nosé, S. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 1984, 52, 255–268. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef] [Green Version]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef]
- Periole, X.; Cavalli, M.; Marrink, S.-J.; Ceruso, M.A. Combining an Elastic Network with a Coarse-Grained Molecular Force Field: Structure, Dynamics, and Intermolecular Recognition. J. Chem. Theory Comput. 2009, 5, 2531–2543. [Google Scholar] [CrossRef] [Green Version]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [Green Version]
- Wassenaar, T.A.; Pluhackova, K.; Böckmann, R.A.; Marrink, S.J.; Tieleman, D.P. Going Backward: A Flexible Geometric Approach to Reverse Transformation from Coarse Grained to Atomistic Models. J. Chem. Theory Comput. 2014, 10, 676–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapadka, K.L.; Becher, F.J.; Dos Santos, A.L.G.; Jackson, S. Factors affecting the physical stability (aggregation) of peptide therapeutics. Interface Focus 2017, 7, 20170030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, V.; Sanchez-Ruiz, J.M. Exploring protein-folding ensembles: A variable-barrier model for the analysis of equilibrium unfolding experiments. Proc. Natl. Acad. Sci. USA 2004, 101, 17646–17651. [Google Scholar] [CrossRef] [Green Version]
- Chebotareva, N.A.; Roman, S.G.; Kurganov, B.I. Dissociative mechanism for irreversible thermal denaturation of oligomeric proteins. Biophys. Rev. 2016, 8, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Monera, O.D.; Kay, C.M.; Hodges, R.S. Protein denaturation with guanidine hydrochloride or urea provides a different estimate of stability depending on the contributions of electrostatic interactions. Protein Sci. 1994, 3, 1984–1991. [Google Scholar] [CrossRef] [Green Version]
- Clayton, K.N.; Salameh, J.W.; Wereley, S.T.; Kinzer-Ursem, T.L. Physical characterization of nanoparticle size and surface modification using particle scattering diffusometry. Biomicrofluidics 2016, 10, 054107. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-Y.; Huang, H.-M.; Lin, C.-C.; Lin, A.F.-Y.; Chan, Y.-C. Effect of Temperature on Hydrophobic Interaction between Proteins and Hydrophobic Adsorbents: Studies by Isothermal Titration Calorimetry and the van’t Hoff Equation. Langmuir 2003, 19, 9395–9403. [Google Scholar] [CrossRef]
- Wang, Y.; Lomakin, A.; Kanai, S.; Alex, R.; Benedek, G.B. Transformation of Oligomers of Lipidated Peptide Induced by Change in pH. Mol. Pharm. 2015, 12, 411–419. [Google Scholar] [CrossRef]
- Wang, X.; Graveland-Bikker, J.F.; De Kruif, C.G.; Robillard, G.T. Oligomerization of hydrophobin SC3 in solution: From soluble state to self-assembly. Protein Sci. 2004, 13, 810–821. [Google Scholar] [CrossRef] [Green Version]
- Diociaiuti, M.; Macchia, G.; Paradisi, S.; Frank, C.; Camerini, S.; Chistolini, P.; Gaudiano, M.C.; Petrucci, T.C.; Malchiodi-Albedi, F. Native metastable prefibrillar oligomers are the most neurotoxic species among amyloid aggregates. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1622–1629. [Google Scholar] [CrossRef] [Green Version]
- Caputo, N.; Jackson, M.A.; Castle, J.R.; El Youssef, J.; Bakhtiani, P.A.; Bergstrom, C.P.; Carroll, J.M.; Breen, M.E.; Leonard, G.L.; David, L.L.; et al. Biochemical Stabilization of Glucagon at Alkaline pH. Diabetes Technol. Ther. 2014, 16, 747–758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, S.; Birkett, N.R.; Fowler, S.B.; Luisi, B.F.; Dobson, C.M.; Zurdo, J. Amyloidogenicity and aggregate cytotoxicity of human glucagon-like peptide-1 (hGLP-1). Protein Pept. Lett. 2009, 16, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, H.; Saito, J.; Tanabe, A.; Amada, T.; Asakura, T.; Kitagawa, K.; Asada, S. Characterization of Novel Insulin Fibrils That Show Strong Cytotoxicity Under Physiological pH. J. Pharm. Sci. 2016, 105, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.; Zhang, C.; Di Domizio, J.; Jin, F.; Connell, W.; Hung, M.; Malkoff, N.; Veksler, V.; Gilliet, M.; Ren, P.; et al. Helical antimicrobial peptides assemble into protofibril scaffolds that present ordered dsDNA to TLR9. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Engelberg, Y.; Landau, M. The Human LL-37(17-29) antimicrobial peptide reveals a functional supramolecular structure. Nat. Commun. 2020, 11, 1–10. [Google Scholar] [CrossRef]
- Hoover, D.M.; Rajashankar, K.R.; Blumenthal, R.; Puri, A.; Oppenheim, J.J.; Chertov, O.; Lubkowski, J. The Structure of Human β-Defensin-2 Shows Evidence of Higher Order Oligomerization. J. Biol. Chem. 2000, 275, 32911–32918. [Google Scholar] [CrossRef] [Green Version]
- Frederiksen, T.M.; Sønderby, P.; Ryberg, L.A.; Harris, P.; Bukrinski, J.T.; Scharff-Poulsen, A.M.; Elf-Lind, M.N.; Peters, G.H. Oligomerization of a Glucagon-like Peptide 1 Analog: Bridging Experiment and Simulations. Biophys. J. 2015, 109, 1202–1213. [Google Scholar] [CrossRef] [Green Version]
- Belfiore, M.; Cariati, I.; Matteucci, A.; Gaddini, L.; Macchia, G.; Fioravanti, R.; Frank, C.; Tancredi, V.; D’Arcangelo, G.; Diociaiuti, M. Calcitonin native prefibrillar oligomers but not monomers induce membrane damage that triggers NMDA-mediated Ca2+-influx, LTP impairment and neurotoxicity. Sci. Rep. 2019, 9, 5144. [Google Scholar] [CrossRef] [Green Version]
- Porcelli, F.; Buck-Koehntop, B.A.; Thennarasu, S.; Ravula, T.; Veglia, G. Structures of the Dimeric and Monomeric Variants of Magainin Antimicrobial Peptides (MSI-78 and MSI-594) in Micelles and Bilayers, Determined by NMR Spectroscopy. Biochemistry 2006, 45, 5793–5799. [Google Scholar] [CrossRef]
- McCaslin, T.G.; Pagba, C.V.; Yohannan, J.; Barry, B.A. Specific metallo-protein interactions and antimicrobial activity in Histatin-5, an intrinsically disordered salivary peptide. Sci. Rep. 2019, 9, 17303. [Google Scholar] [CrossRef] [Green Version]
- Sancho-Vaello, E.; François, P.; Bonetti, E.-J.; Lilie, H.; Finger, S.; Gil-Ortiz, F.; Gil-Carton, D.; Zeth, K. Structural remodeling and oligomerization of human cathelicidin on membranes suggest fibril-like structures as active species. Sci. Rep. 2017, 7, 15371. [Google Scholar] [CrossRef] [PubMed]
- Egabizon, R.; Friedler, A. Allosteric modulation of protein oligomerization: An emerging approach to drug design. Front. Chem. 2014, 2, 9. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, K.M.; Baldwin, R.L. Charged histidine affects alpha-helix stability at all positions in the helix by interacting with the backbone charges. Proc. Natl. Acad. Sci. USA 1993, 90, 11337–11340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holdbrook, D.A.; Singh, S.; Choong, Y.K.; Petrlova, J.; Malmsten, M.; Bond, P.J.; Verma, N.K.; Schmidtchen, A.; Saravanan, R. Influence of pH on the activity of thrombin-derived antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2374–2384. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Maertens, G.; Cherepanov, P.; Debyser, Z.; Engelborghs, Y.; Engelman, A.N.; Tsumoto, K.; Yasutake, Y.; Umetsu, M.; Yao, M.; et al. How Oligomerization Contributes to the Thermostability of an Archaeon Protein: PROTEIN L-ISOASPARTYL-O-METHYLTRANSFERASE FROM SULFOLOBUS TOKODAII. J. Biol. Chem. 2004, 279, 32957–32967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, H.; Ellington, A.D. Increasing the thermal stability of an oligomeric protein, beta-glucuronidase. J. Mol. Biol. 2002, 315, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Panchenko, A.R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. USA 2010, 107, 20352–20357. [Google Scholar] [CrossRef] [Green Version]
- Pertinhez, T.A.; Conti, S.; Ferrari, E.; Magliani, W.; Spisni, A.; Polonelli, L. Reversible Self-Assembly: A Key Feature for a New Class of Autodelivering Therapeutic Peptides. Mol. Pharm. 2009, 6, 1036–1039. [Google Scholar] [CrossRef]
- Gupta, S.; Chattopadhyay, T.; Singh, M.P.; Surolia, A. Supramolecular insulin assembly II for a sustained treatment of type 1 diabetes mellitus. Proc. Natl. Acad. Sci. USA 2010, 107, 13246–13251. [Google Scholar] [CrossRef] [Green Version]
- Koike, R.; Kidera, A.; Ota, M. Alteration of oligomeric state and domain architecture is essential for functional transformation between transferase and hydrolase with the same scaffold. Protein Sci. 2009, 18, 2060–2066. [Google Scholar] [CrossRef] [Green Version]
- Xhindoli, D.; Pacor, S.; Guida, F.; Antcheva, N.; Tossi, A. Native oligomerization determines the mode of action and biological activities of human cathelicidin LL-37. Biochem. J. 2013, 457, 263–275. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petruk, G.; Petrlova, J.; Samsudin, F.; Giudice, R.D.; Bond, P.J.; Schmidtchen, A. Concentration- and pH-Dependent Oligomerization of the Thrombin-Derived C-Terminal Peptide TCP-25. Biomolecules 2020, 10, 1572. https://doi.org/10.3390/biom10111572
Petruk G, Petrlova J, Samsudin F, Giudice RD, Bond PJ, Schmidtchen A. Concentration- and pH-Dependent Oligomerization of the Thrombin-Derived C-Terminal Peptide TCP-25. Biomolecules. 2020; 10(11):1572. https://doi.org/10.3390/biom10111572
Chicago/Turabian StylePetruk, Ganna, Jitka Petrlova, Firdaus Samsudin, Rita Del Giudice, Peter J. Bond, and Artur Schmidtchen. 2020. "Concentration- and pH-Dependent Oligomerization of the Thrombin-Derived C-Terminal Peptide TCP-25" Biomolecules 10, no. 11: 1572. https://doi.org/10.3390/biom10111572
APA StylePetruk, G., Petrlova, J., Samsudin, F., Giudice, R. D., Bond, P. J., & Schmidtchen, A. (2020). Concentration- and pH-Dependent Oligomerization of the Thrombin-Derived C-Terminal Peptide TCP-25. Biomolecules, 10(11), 1572. https://doi.org/10.3390/biom10111572