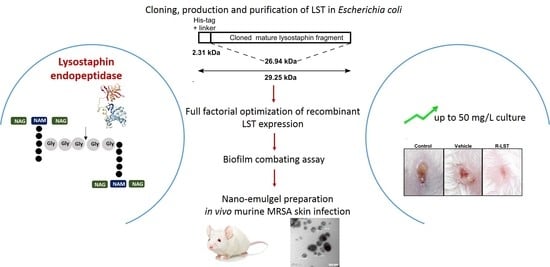
A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform
Abstract
:1. Introduction
2. Materials and Methods
2.1. Statement of Ethical Approval
2.2. Bacterial Strains and Culture Conditions
2.3. Cloning and Expression of Recombinant Lysostaphin in E. coli
2.4. Optimization of Recombinant Lysostaphin Production
2.5. Analyses of the Different Factors and Their Interactions on the Production of rLST
2.6. Lysostaphin Activity Assay
2.7. Biofilms Combating Assays
2.7.1. Determination of the Minimum Inhibitory Concentration (MIC)
2.7.2. Biofilm Formation Inhibition Assay (Pre-Exposure)
2.7.3. Biofilm Detachment Assay (Post-Exposure)
2.8. Incorporation of the rLST into Nano-Emulgel Preparation
2.8.1. Characterization of the rLST Nano-Emulgel
2.8.2. In-Vitro Release Study
2.9. Testing the Efficacy of the rLST Nano-Emulgel in an In-Vivo Murine Skin Model of Infection
3. Results
3.1. Cloning and Recombinant Lysostaphin Production
3.2. Optimization of rLST Production
3.3. Laboratory-Scale Production Cost of rLST
3.4. The rLST Is Active in Lysing Staphylococcal Cells
3.5. The rLST Is Active in Inhibiting and Combating Staphylococcal Biofilms
3.6. The rLST Is Successfully Incorporated into a Nano-Emulgel and Released While Retaining Its Activity
3.7. The rLST Is Active in Curing S. aureus Skin Infection in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- W.H.O. Prioritization of Pathogens to Guide Discovery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Savoia, D. Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol. 2012, 7, 979–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burrowes, B.; Harper, D.R.; Anderson, J.; McConville, M.; Enright, M.C. Bacteriophage therapy: potential uses in the control of antibiotic-resistant pathogens. Expert Rev. Anti-Infect. Ther. 2011, 9, 775–785. [Google Scholar] [CrossRef]
- Kole, R.; Krainer, A.R.; Altman, S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 2012, 11, 125–140. [Google Scholar] [CrossRef] [Green Version]
- El-Halfawy, O.M.; Czarny, T.L.; Flannagan, R.S.; Day, J.; Bozelli, J.C.J.; Kuiack, R.C.; Salim, A.; Eckert, P.; Epand, R.M.; McGavin, M.J.; et al. Discovery of an antivirulence compound that reverses beta-lactam resistance in MRSA. Nat. Chem. Biol. 2019, 16, 143–149. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, Q.; Zhang, Q.; Huang, Y.; Su, Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides 2012, 37, 207–215. [Google Scholar] [CrossRef]
- Bannerman, T.L. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically. Man. Clin. Microbiol. 2003, 8, 384–404. [Google Scholar]
- do Carmo de Freire Bastos, M.; Coutinho, B.G.; Coelho, M.L.V. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals 2010, 3, 1139–1161. [Google Scholar] [CrossRef] [Green Version]
- Koroleva, M.Y.; Yurtov, E.V. Nanoemulsions: the properties, methods of preparation and promising applications. Russ. Chem. Rev. 2012, 81, 21. [Google Scholar] [CrossRef]
- Azmi, N.A.N.; Elgharbawy, A.A.M.; Motlagh, S.R.; Samsudin, N.; Salleh, H.M. Nanoemulsions: Factory for Food, Pharmaceutical and Cosmetics. Processes 2019, 7, 617. [Google Scholar] [CrossRef] [Green Version]
- Ojha, A.; Ojha, M.; Madhav, N.V.S. Recent Advancement in Emulgel: A Novel Approach for Topical Drug Delivery. Int. J. Adv. Pharm. 2017, 6, 1–21. [Google Scholar]
- Patel, Z.S.; Shah, R.; Shah, D.N. Nanoemulsion: An innovative approach for topical delivery. Pharma Sci. Monit. 2016, 7, 21–36. [Google Scholar]
- Salome Amarachi, C.; Kenechukwu, F.; Attama, A. Nanoemulsions — Advances in formulation, characterization and applications in drug delivery. In Application of Nanotechnology in Drug Delivery; Sezer, A.D., Ed.; IntechOpen: London, UK, 2014; pp. 76–126. [Google Scholar]
- Lorenz, L.L.; Duthie, E.S. Staphylococcal Coagulase: Mode of Action and Antigenicity. Microbiology 1952, 6, 95–107. [Google Scholar]
- Diep, B.A.; Gill, S.R.; Chang, R.F.; Phan, T.H.; Chen, J.H.; Davidson, M.G.; Lin, F.; Lin, J.; Carleton, H.A.; Mongodin, E.F.; et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 2006, 367, 731–739. [Google Scholar] [CrossRef]
- Daniel, A.; Euler, C.; Collin, M.; Chahales, P.; Gorelick, K.J.; Fischetti, V.A. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 1603–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heilbronner, S.; Holden, M.T.G.; van Tonder, A.; Geoghegan, J.A.; Foster, T.J.; Parkhill, J.; Bentley, S.D. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors. FEMS Microbiol. Lett. 2011, 322, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Recsei, P.A.; Gruss, A.D.; Novick, R.P. Cloning, sequence, and expression of the lysostaphin gene from Staphylococcus simulans. Proc. Natl. Acad. Sci. USA 1987, 84, 1127–1131. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, D.P.; Nanavaty, T.; McDevitt, D.; Gurusiddappa, S.; Höök, M.; Foster, T.J. The Fibrinogen-binding MSCRAMM (Clumping Factor) of Staphylococcus aureus has a Ca 2+ -dependent inhibitory site. J. Biol. Chem. 1998, 273, 6821–6829. [Google Scholar] [CrossRef] [Green Version]
- Szweda, P.; Kotłowski, R.; Kur, J. New effective sources of the Staphylococcus simulans lysostaphin. J. Biotechnol. 2005, 117, 203–213. [Google Scholar] [CrossRef] [PubMed]
- CLSI. M100 Performance Standards for Antimicrobial Susceptibility Testing an Informational Supplement for Global Application Developed through the Clinical and Laboratory Standards Institute Consensus Process; CLSI: Wayne, PA, USA, 2016; ISBN 1562388045. [Google Scholar]
- O’Toole, G.A. Microtiter dish biofilm formation assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Yuan, F.; Lu, F.; Yin, Y.; Cao, J. Vancomycin-induced biofilm formation by methicillin-resistant Staphylococcus aureus is associated with the secretion of membrane vesicles. Microb. Pathog. 2017, 110, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.A.; Corrigan, R.M.; Gruszka, D.T.; Speziale, P.; O’Gara, J.P.; Potts, J.R.; Foster, T.J. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 2010, 192, 5663–5673. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.A.; Kusuma, C.; Mond, J.J.; Kokai-Kun, J.F. Lysostaphin disrupts Staphylococcus aureus and Staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob. Agents Chemother. 2003, 47, 3407–3414. [Google Scholar] [CrossRef] [Green Version]
- Forgiarini, A.; Esquena, J.; González, C.; Solans, C. Formation of nano-emulsions by low-energy emulsification methods at constant temperature. Langmuir 2001, 17, 2076–2083. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Hamed, M.I.; Panitch, A.; Seleem, M.N. Targeting methicillin-resistant Staphylococcus aureus with short salt-resistant synthetic peptides. Antimicrob. Agents Chemother. 2014, 58, 4113–4122. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, M.F.; Abdelkhalek, A.; Seleem, M.N. Evaluation of short synthetic antimicrobial peptides for treatment of drug-resistant and intracellular Staphylococcus aureus. Sci. Rep. 2016, 6, 29707. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: New York, NA, USA, 2005; pp. 571–607. [Google Scholar]
- Sharma, R.; Sharma, P.R.; Choudhary, M.L.; Pande, A.; Khatri, G.S. Cytoplasmic expression of mature glycylglycine endopeptidase lysostaphin with an amino terminal hexa-histidine in a soluble and catalytically active form in Escherichia coli. Protein Expr Purif. 2006, 45, 206–215. [Google Scholar] [CrossRef]
- Zhang, B.; Shangguan, T.; Ma, H.; Huang, X.; Zhang, Y. Lysis of mastitis pathogens isolated from dairy cow milk samples by purified recombinant lysostaphin. Afr. J. Biotechnol. 2012, 11, 4649–4659. [Google Scholar]
- Farhangnia, L.; Ghaznavi-Rad, E.; Mollaee, N.; Abtahi, H. Cloning, expression, and purification of recombinant lysostaphin from staphylococcus simulans. Jundishapur J. Microb. 2014, 7, e10009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szweda, P.; Gorczyca, G.; Filipkowski, P.; Zalewska, M.; Milewski, S. Efficient production of Staphylococcus simulans lysostaphin in a benchtop bioreactor by recombinant Escherichia coli. Prep. Biochem. Biotech. 2014, 44, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Kiedrowski, M.R.; Horswill, A.R. New approaches for treating staphylococcal biofilm infections. Ann. N. Y. Acad. Sci. 2011, 1241, 104–121. [Google Scholar] [CrossRef]
- Rasmussen, T.B.; Givskov, M. Quorum-sensing inhibitors as anti-pathogenic drugs. Int.J. Med. Microbiol. 2006, 296, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Antibiotic-induced biofilm formation. Int. J. Artif. Organs. 2011, 34, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Mirani, Z.A.; Jamil, N. Effect of sub-lethal doses of vancomycin and oxacillin on biofilm formation by vancomycin intermediate resistant Staphylococcus aureus. J. Basic Microbiol. 2011, 51, 191–195. [Google Scholar] [CrossRef]
- Frank, K.L.; Reichert, E.J.; Piper, K.E.; Patel, R. In vitro effects of antimicrobial agents on planktonic and biofilm forms of Staphylococcus lugdunensis clinical isolates. Antimicrob. Agents Chemother. 2007, 51, 888–895. [Google Scholar] [CrossRef] [Green Version]
- Stranden, A.M.; Ehlert, K.; Labischinski, H.; Berger-Bächi, B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 1997, 179, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Kusuma, C.; Jadanova, A.; Chanturiya, T.; Kokai-Kun, J.F. Lysostaphin-resistant variants of Staphylococcus aureus demonstrate reduced fitness in vitro and in vivo. Antimicrob. agents Chemother. 2007, 51, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Harrison, E.; Cropp, C. Therapeutic activity of lysostaphin in experimental staphylococcal infections. Can. J. Microbiol. 2011, 13, 93–97. [Google Scholar] [CrossRef]
- Climo, M.W.; Patron, R.L.; Goldstein, B.P.; Archer, G.L. Lysostaphin treatment of experimental methicillin-resistant Staphylococcus aureus aortic valve endocarditis. Antimicrob. Agents Chemother. 1998, 42, 1355–1360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiri, N.; Archer, G.; Climo, M.W. Combinations of lysostaphin with beta-lactams are synergistic against oxacillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 2002, 46, 2017–2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Placencia, F.X.; Kong, L.; Weisman, L.E. Treatment of methicillin-resistant Staphylococcus aureus in neonatal mice: lysostaphin versus vancomycin. Pediatr. Res. 2009, 65, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, V.; Ravichandran, V. Nanoemulsions: As modified drug delivery tool advantages of nanoemulsion. Int. J. Compr. Pharm. 2011, 2, 1–6. [Google Scholar]
- Schindler, C.; Schuhardt, V. Lysostaphin: A new bacteriolytic agent for the staphylococcus. Proc. Natl. Acad. Sci. USA 1964, 51, 414–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbois, A.P.; Lang, S.; Gemmell, C.G.; Coote, P.J. Surface disinfection properties of the combination of an antimicrobial peptide, ranalexin, with an endopeptidase, lysostaphin, against methicillin-resistant Staphylococcus aureus (MRSA). J. Appl. Microbiol. 2010, 108, 723–730. [Google Scholar] [CrossRef]
- Mohamed, M.F.; Seleem, M.N. Efficacy of short novel antimicrobial and anti-inflammatory peptides in a mouse model of methicillin-resistant Staphylococcus aureus (MRSA) skin infection. Drug Des. Devel. Ther. 2014, 8, 1979–1983. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Li, G.; Huang, J.; Zhang, J.; Lu, M.; Lu, W.; Huang, Q. Extension of nasal anti-Staphylococcus aureus efficacy of lysostaphin by its incorporation into a chitosan-o/w cream. Drug Deliv. 2010, 17, 617–623. [Google Scholar] [CrossRef]






© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nour El-Din, H.T.; Elhosseiny, N.M.; El-Gendy, M.A.; Mahmoud, A.A.; Hussein, M.M.M.; Attia, A.S. A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform. Biomolecules 2020, 10, 435. https://doi.org/10.3390/biom10030435
Nour El-Din HT, Elhosseiny NM, El-Gendy MA, Mahmoud AA, Hussein MMM, Attia AS. A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform. Biomolecules. 2020; 10(3):435. https://doi.org/10.3390/biom10030435
Chicago/Turabian StyleNour El-Din, Hanzada T., Noha M. Elhosseiny, Mohamed A. El-Gendy, Azza A. Mahmoud, Manal M.M. Hussein, and Ahmed S. Attia. 2020. "A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform" Biomolecules 10, no. 3: 435. https://doi.org/10.3390/biom10030435
APA StyleNour El-Din, H. T., Elhosseiny, N. M., El-Gendy, M. A., Mahmoud, A. A., Hussein, M. M. M., & Attia, A. S. (2020). A Rapid Lysostaphin Production Approach and a Convenient Novel Lysostaphin Loaded Nano-emulgel; As a Sustainable Low-Cost Methicillin-Resistant Staphylococcus aureus Combating Platform. Biomolecules, 10(3), 435. https://doi.org/10.3390/biom10030435