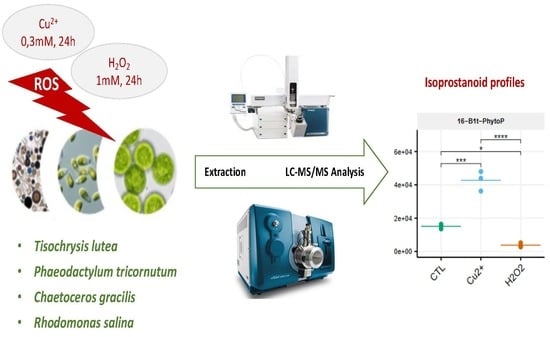
Isoprostanoid Profiling of Marine Microalgae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Microalgal Species
2.3. Cultivation of Microalgae and Oxidative Stress Treatments
2.4. Preparation of Algal Samples for Lipidomic Analysis
2.5. Preparation of Samples for Analysis of Extraction Yield and Matrix Effect
2.6. Micro-LC-MS/MS Analysis
2.7. Statistical Analysis
3. Results
3.1. Analysis of Extraction Yield and Matrix Effect
3.2. Rhodomonas Salina
3.3. Tisochrysis Lutea
3.4. Chaetoceros Gracilis
3.5. Phaeodactylum Tricornutum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Worden, A.Z.; Follows, M.J.; Giovannoni, S.J.; Wilken, S.; Zimmerman, A.E.; Keeling, P.J. Environmental science. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 2015, 347, 1257594. [Google Scholar] [CrossRef] [Green Version]
- Ebenezer, V.; Medlin, L.K.; Ki, J.S. Molecular detection, quantification, and diversity evaluation of microalgae. Mar. Biotechnol. 2012, 14, 129–142. [Google Scholar] [CrossRef]
- Brodie, J.; Chan, C.X.; de Clerck, O.; Cock, J.M.; Coelho, S.M.; Gachon, C.; Grossman, A.R.; Mock, T.; Raven, J.A.; Smith, A.G.; et al. The Algal Revolution. Trends Plant Sci. 2017, 22, 726–738. [Google Scholar] [CrossRef]
- Venkata, M.S.; Hemalatha, M.; Chakraborty, D.; Chatterjee, S.; Ranadheer, P.; Kona, R. Algal biorefinery models with self-sustainable closed loop approach: Trends and prospective for blue-bioeconomy. Bioresour. Technol. 2020, 295, 122128. [Google Scholar] [CrossRef] [PubMed]
- Castro, L.F.; Tocher, D.R.; Monroig, O. Long-chain polyunsaturated fatty acid biosynthesis in chordates: Insights into the evolution of Fads and Elovl gene repertoire. Prog. Lipid Res. 2016, 62, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Very long chain omega-3 (n-3) fatty acids and human health. Eur. J. Lipid Sci. Technol. 2014, 116, 1280–1300. [Google Scholar] [CrossRef]
- Calder, P.C. Very long-chain n-3 fatty acids and human health: Fact, fiction and the future. Proc. Nutr. Soc. 2018, 77, 52–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Lista, J.; Perez-Martinez, P.; Lopez-Miranda, J.; Perez-Jimenez, F. Long chain omega-3 fatty acids and cardiovascular disease: A systematic review. Br. J. Nutr. 2012, 107, S201–S213. [Google Scholar] [CrossRef] [Green Version]
- Innis, S.M. Dietary (n-3) fatty acids and brain development. J. Nutr. 2007, 137, 855–859. [Google Scholar] [CrossRef] [Green Version]
- Campoy, C.; Escolano-Margarit, M.V.; Anjos, T.; Szajewska, H.; Uauy, R. Omega 3 fatty acids on child growth, visual acuity and neurodevelopment. Br. J. Nutr. 2012, 10, S85–S106. [Google Scholar] [CrossRef]
- Hilgendorf, K.I.; Johnson, C.T.; Mezger, A.; Rice, S.L.; Norris, A.M.; Demeter, J.; Greenleaf, W.J.; Reiter, J.F.; Kopinke, D.; Jackson, P.K. Omega-3 Fatty Acids Activate Ciliary FFAR4 to Control Adipogenesis. Cell 2019, 179, 1289–1305. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R.; Betancor, M.B.; Sprague, M.; Olsen, R.E.; Napier, J.A. Omega-3 Long-Chain Polyunsaturated Fatty Acids, EPA and DHA: Bridging the Gap between Supply and Demand. Nutrients 2019, 11, 89. [Google Scholar] [CrossRef] [Green Version]
- Diao, J.; Song, X.; Guo, T.; Wang, F.; Chen, L.; Zhang, W. Cellular engineering strategies toward sustainable omega-3 long chain polyunsaturated fatty acids production: State of the art and perspectives. Biotechnol. Adv. 2020, 40, 107497. [Google Scholar] [CrossRef] [PubMed]
- Napier, J.A.; Olsen, R.-E.; Tocher, D.R. Update on GM canola crops as novel sources of omega-3 fish oils. Plant. Biotechnol. J. 2019, 17, 703–705. [Google Scholar] [CrossRef] [PubMed]
- Jahn, U.; Galano, J.-M.; Durand, T. Beyond Prostaglandins—Chemistry and Biology of Cyclic Oxygenated Metabolites Formed by Free-Radical Pathways from Polyunsaturated Fatty Acids. Angew. Chem. Int. Ed. 2008, 47, 5894–5955. [Google Scholar] [CrossRef]
- Galano, J.M.; Lee, Y.Y.; Oger, C.; Vigor, C.; Vercauteren, J.; Durand, T.; Giera, M.; Lee, J.C. Isoprostanes, neuroprostanes and phytoprostanes: An overview of 25years of research in chemistry and biology. Prog. Lipid Res. 2017, 68, 83–108. [Google Scholar] [CrossRef]
- Barbosa, M.; Collado-Gonzalez, J.; Andrade, P.B.; Ferreres, F.; Valentao, P.; Galano, J.M.; Durand, T.; Gil-Izquierdo, A. Nonenzymatic alpha-Linolenic Acid Derivatives from the Sea: Macroalgae as Novel Sources of Phytoprostanes. J. Agric. Food Chem. 2015, 63, 6466–6474. [Google Scholar] [CrossRef]
- Vigor, C.; Reversat, G.; Rocher, A.; Oger, C.; Galano, J.M.; Vercauteren, J.; Durand, T.; Tonon, T.; Leblanc, C.; Potin, P. Isoprostanoids quantitative profiling of marine red and brown macroalgae. Food Chem. 2018, 268, 452–462. [Google Scholar] [CrossRef] [Green Version]
- Lupette, J.; Jaussaud, A.; Vigor, C.; Oger, C.; Galano, J.M.; Reversat, G.; Vercauteren, J.; Jouhet, J.; Durand, T.; Marechal, E. Non-enzymatic synthesis of bioactive isoprostanoids in the diatom Phaeodactylum following oxidative stress. Plant. Physiol. 2018, 178, 1344–1357. [Google Scholar] [CrossRef] [Green Version]
- Kovac, D.J.; Simeunovic, J.B.; Babic, O.B.; Misan, A.C. Algae in food and feed. Food Feed Res. 2013, 40, 21–32. [Google Scholar]
- Chauton, M.S.; Reitan, K.I.; Norsker, N.H.; Tveterås, R.; Kleivdal, H.T. A techno-economic analysis of industrial production of marine microalgae as a source of EPA and DHA-rich raw material for aquafeed: Research challenges and possibilities. Aquaculture 2015, 436, 95–103. [Google Scholar] [CrossRef]
- Kumari, P.; Kumar, M.; Reddy, C.R.K.; Jha, B. Algal lipids, fatty acids and sterols. In Functional Ingredients from Algae for Foods and Nutraceuticals; Dominguez, H., Ed.; Woodhead: Cambridge, UK, 2013; pp. 87–134. [Google Scholar]
- Oger, C.; Brinkmann, Y.; Bouazzaoui, S.; Durand, T.; Galano, J.M. Stereocontrolled access to isoprostanes via a bicyclo[3.3.0]octene framework. Org. Lett. 2008, 10, 5087–5090. [Google Scholar] [CrossRef] [PubMed]
- Oger, C.; Bultel-Ponce, V.; Guy, A.; Durand, T.; Galano, J.M. Total synthesis of isoprostanes derived from AdA and EPA. Eur. J. Org. Chem. 2012, 2012, 2621–2634. [Google Scholar] [CrossRef]
- Guy, A.; Oger, C.; Heppekausen, J.; Signorini, C.; de Felice, C.; Fürstner, A.; Durand, T.; Galano, J. Oxygenated metabolites of n-3 polyunsaturated fatty acids as potential oxidative stress biomarkers: Total synthesis of 8-F3t-IsoP, 10-F4t-NeuroP and [D4]-10-F4t-NeuroP. Chem. Eur. J. 2014, 20, 6374–6380. [Google Scholar] [CrossRef]
- Cuyamendous, C.; Leung, K.S.; Durand, T.; Lee, J.C.; Oger, C.; Galano, J.M. Synthesis and discovery of phytofurans: Metabolites of alpha-linolenic acid peroxidation. Chem. Commun. 2015, 51, 15696–15699. [Google Scholar] [CrossRef]
- Lananan, F.; Jusoh, A.; Ali, N.; Lam, S.S.; Endut, A. Effect of Conway Medium and f/2 Medium on the growth of six genera of South China Sea marine microalgae. Bioresour. Technol. 2013, 141, 75–82. [Google Scholar] [CrossRef]
- Heydarizadeh, P.; Boureba, W.; Zahedi, M.; Huang, B.; Moreau, B.; Lukomska, E.; Couzinet-Mossion, A.; Wielgosz-Collin, G.; Martin-Jézéquel, V.; Bougaran, G.; et al. Response of CO2-starved diatom Phaeodactylum tricornutum to light intensity transition. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160396. [Google Scholar] [CrossRef] [Green Version]
- del Pilar Sánchez-Saavedra, M.; Maeda-Martínez, A.N.; Acosta-Galindo, S. Effect of different light spectra on the growth and biochemical composition of Tisochrysis lutea. J. Appl. Phycol. 2016, 28, 839–847. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, R version 3.5.4 Patched; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 11 March 2019).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; D’Agostino McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the tidyverse. J. Open Source Softw. 2019, 4. [Google Scholar] [CrossRef]
- Zhu, H.; Travison, T.; Tsai, T.; Beasley, W.; Xie, Y.; Yu, G.; Laurent, S.; Shepherd, R.; Sidi, Y.; Salzer, B.G.; et al. KableExtra: Construct Complex Table with ‘kable’ and Pipe Syntax. R package version 1.1.0. Available online: https://CRAN.R-project.org/package=kableExtra (accessed on 1 January 2019).
- Kassambara, K. Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. R package version 0.4.0. Available online: https://CRAN.R-project.org/package=rstatix (accessed on 1 January 2020).
- Leung, K.S.; Chen, X.; Zhong, W.; Yu, A.C.; Lee, C.Y. Microbubble-mediated sonoporation amplified lipid peroxidation of Jurkat cells. Chem. Phys. Lipids 2014, 180, 53–60. [Google Scholar] [CrossRef]
- Dupuy, A.; Le Faouder, P.; Vigor, C.; Oger, C.; Galano, J.M.; Dray, C.; Lee, J.C.; Valet, P.; Gladine, C.; Durand, T.; et al. Simultaneous quantitative profiling of 20 isoprostanoids from omega-3 and omega-6 polyunsaturated fatty acids by LC-MS/MS in various biological samples. Anal. Chim. Acta 2016, 921, 46–58. [Google Scholar] [CrossRef] [Green Version]
- Yonny, M.E.; Rodriguez Torresi, A.; Cuyamendous, C.; Reversat, G.; Oger, C.; Galano, J.M.; Durand, T.; Vigor, C.; Nazareno, M.A. Thermal Stress in Melon Plants: Phytoprostanes and Phytofurans as Oxidative Stress Biomarkers and the Effect of Antioxidant Supplementation. J. Agric. Food Chem. 2016, 64, 8296–8304. [Google Scholar] [CrossRef]
- Ruesgas-Ramón, M.; Figueroa-Espinoza, M.; Durand, E.; Suárez-Quiroz, M.; González-Ríos, O.; Rocher, A.; Reversat, G.; Vercauteren, J.; Oger, C.; Galano, J.; et al. Identification and quantification of phytoprostanes and phytofurans of coffee and cocoa by- and co-products. Food Funct. 2019, 10, 6882–6891. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Roldan, N.; Engel, R.; Dupow, S.; Jakob, K.; Koops, F.; Orinska, Z.; Vigor, C.; Oger, C.; Galano, J.M.; Durand, T.; et al. Lipid Mediators From Timothy Grass Pollen Contribute to the Effector Phase of Allergy and Prime Dendritic Cells for Glycolipid Presentation. Front. Immunol. 2019, 10, 974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jónasdóttir, S.H. Fatty Acid Profiles and Production in Marine Phytoplankton. Mar. Drugs 2019, 17, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Reiriz, M.J.; Perez-Camacho, A.; Ferreiro, M.J.; Blanco, J.; Planas, M.; Campos, M.J.; Labarta, U. Biomass production and variation in the biochemical profile (total protein, carbohydrates, RNA, lipids and fatty acids) of seven species of marine microalgae. Aquaculture 1989, 83, 17–37. [Google Scholar] [CrossRef]
- Guevara, M.; Bastardo L Fau-Cortez, R.; Cortez R Fau-Arredondo-Vega, B.; Arredondo-Vega B Fau-Romero, L.; Romero L Fau-Gómez, P.; Gómez, P. Rhodomonas salina (Cryptophyta) pastes as feed for Brachionus plicatilis (Rotifera). Rev. Biol. Trop. 2011, 59, 1503–1515. [Google Scholar]
- Huang, B.; Marchand, J.; Thiriet-Rupert, S.; Carrier, G.; Saint-Jean, B.; Lukomska, E.; Moreau, B.; Morant-Manceau, A.; Bougaran, G.; Mimouni, V. Betaine lipid and neutral lipid production under nitrogen or phosphorus limitation in the marine microalga Tisochrysis lutea (Haptophyta). Algal Res. 2019, 40, 101506. [Google Scholar] [CrossRef]
- Volkman, J.K.; Jeffrey, S.W.; Nichols, P.D.; Rogers, G.I.; Garland, C.D. Fatty acid and lipid composition of 10 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 1989, 128, 219–240. [Google Scholar] [CrossRef]
- Pratiwi, A.R.; Syah, D.; Hardjito, L.; Panggabean, L.M.G.; Suhartono, M.T. Fatty Acid Synthesis by Indonesian Marine Diatom, Chaetoceros gracilis. HAYATI J. Biosci. 2009, 16, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Thomas-Hall, S.R.; Schenk, P.M. Phaeodactylum tricornutum microalgae as a rich source of omega-3 oil: Progress in lipid induction techniques towards industry adoption. Food Chem. 2019, 297, 124937. [Google Scholar] [CrossRef] [PubMed]
- Di Dato, V.; Orefice, I.; Amato, A.; Fontanarosa, C.; Amoresano, A.; Cutignano, A.; Ianora, A.; Romano, G. Animal-like prostaglandins in marine microalgae. ISME J. 2017, 11, 1722–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Dato, V.; Barbarinaldi, R.; Amato, A.; Di Costanzo, F.; Fontanarosa, C.; Perna, A.; Amoresano, A.; Esposito, F.; Cutignano, A.; Lanora, A.; et al. Variation in prostaglandin metabolism during growth of the diatom Thalassiosira rotula. Sci. Rep. 2020, 10, 5374. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Rettner, J.; Werner, M.; Werz, O.; Pohnert, G. Algal Oxylipins Mediate the Resistance of Diatoms against Algicidal Bacteria. Mar. Drugs 2018, 16, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russo, E.; d’Ippolito, G.; Fontana, A.; Sarno, D.; D’Alelio, D.; Busseni, G.; Ianora, A.; von Elert, E.; Carotenuto, Y. Density-dependent oxylipin production in natural diatom communities: Possible implications for plankton dynamics. ISME J. 2020, 14, 164–177. [Google Scholar] [CrossRef]
- He, Q.; Yang, H.; Wu, L.; Hu, C. Effect of light intensity on physiological changes, carbon allocation and neutral lipid accumulation in oleaginous microalgae. Bioresour. Technol. 2015, 191, 219–228. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Chen, H.; He, C.-L.; Wang, Q. Nitrogen Starvation Induced Oxidative Stress in an Oil-Producing Green Alga Chlorella sorokiniana C3. PLoS ONE 2013, 8, e69225. [Google Scholar] [CrossRef]
- Yilancioglu, K.; Cokol, M.; Pastirmaci, I.; Erman, B.; Cetiner, S. Oxidative Stress Is a Mediator for Increased Lipid Accumulation in a Newly Isolated Dunaliella salina Strain. PLoS ONE 2014, 9, e91957. [Google Scholar] [CrossRef]
- Renaud, S.M.; Thinh, L.-V.; Lambrinidis, G.; Parry, D.L. Effect of temperature on growth, chemical composition and fatty acid composition of tropical Australian microalgae grown in batch cultures. Aquaculture 2002, 211, 195–214. [Google Scholar] [CrossRef]
- Church, J.; Hwang, J.H.; Kim, K.T.; McLean, R.; Oh, Y.K.; Nam, B.; Joo, J.C.; Lee, W.H. Effect of salt type and concentration on the growth and lipid content of Chlorella vulgaris in synthetic saline wastewater for biofuel production. Bioresour. Technol. 2017, 243, 147–153. [Google Scholar] [CrossRef]
- Watrous, J.D.; Niiranen, T.J.; Lagerborg, K.A.; Henglin, M.; Xu, Y.-J.; Rong, J.; Sharma, S.; Vasan, R.S.; Larson, M.G.; Armando, A.; et al. Directed Non-targeted Mass Spectrometry and Chemical Networking for Discovery of Eicosanoids and Related Oxylipins. Cell Chem. Biol. 2019, 26, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Martínez, E.; Cosnahan, R.K.; Wu, M.; Gadila, S.K.; Quick, E.B.; Mobley, J.A.; Campos-Gómez, J. Oxylipins mediate cell-to-cell communication in Pseudomonas aeruginosa. Commun. Biol. 2019, 2, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, H.; Johnson, D.; Gensler, C.; Decker, E.; Zhang, G. Chemistry and biology of ω-3 PUFA peroxidation-derived compounds. Prostag. Other Lipid Mediat. 2017, 132, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Minghetti, L.; Salvi, R.; Lavinia Salvatori, M.; Antonietta Ajmone-Cat, M.; De Nuccio, C.; Visentin, S.; Bultel-Poncé, V.; Oger, C.; Guy, A.; Galano, J.-M.; et al. Nonenzymatic oxygenated metabolites of α-linolenic acid B1- and L1-phytoprostanes protect immature neurons from oxidant injury and promote differentiation of oligodendrocyte progenitors through PPAR-γ activation. Free Radic. Biol. Med. 2014, 73, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.Y.; Galano, J.-M.; Leung, H.H.; Balas, L.; Oger, C.; Durand, T.; Lee, J.C.-Y. Nonenzymatic oxygenated metabolite of docosahexaenoic acid, 4(RS)-4-F4t-neuroprostane, acts as a bioactive lipid molecule in neuronal cells. FEBS Lett. 2020, 594, 1797–1808. [Google Scholar] [CrossRef]
- Song, W.L.; Paschos, G.; Fries, S.; Reilly, M.P.; Yu, Y.; Rokach, J.; Chang, C.T.; Patel, P.; Lawson, J.A.; Fitzgerald, G.A. Novel eicosapentaenoic acid-derived F3-isoprostanes as biomarkers of lipid peroxidation. J. Biol. Chem. 2009, 284, 23636–23643. [Google Scholar] [CrossRef] [Green Version]
- Praticò, D.; Smyth Em Fau-Violi, F.; Violi F Fau-FitzGerald, G.A.; FitzGerald, G.A. Local amplification of platelet function by 8-Epi prostaglandin F2alpha is not mediated by thromboxane receptor isoforms. J. Biol. Chem. 1996, 271, 14916–14924. [Google Scholar] [CrossRef] [Green Version]
- Roy, J.; Oger, C.; Thireau, J.; Roussel, J.; Mercier-Touzet, O.; Faure, D.; Pinot, E.; Farah, C.; Taber, D.F.; Cristol, J.-P.; et al. Guennec, Nonenzymatic lipid mediators, neuroprostanes, exert the antiarrhythmic properties of docosahexaenoic acid. Free Radic. Biol. Med. 2015, 86, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Roy, J.; Fauconnier, J.; Oger, C.; Farah, C.; Angebault-Prouteau, C.; Thireau, J.; Bideaux, P.; Scheuermann, V.; Bultel-Poncé, V.; Demion, M.; et al. Guennec, Non-enzymatic oxidized metabolite of DHA, 4(RS)-4-F(4t)-neuroprostane protects the heart against reperfusion injury. Free Radic. Biol. Med. 2017, 102, 229–239. [Google Scholar] [CrossRef]






| Microalgal Species | Metabolites of ALA | Metabolites of AA | Metabolites of AdA | Metabolites of EPA | Metabolites of DPA | Metabolites of DHA |
|---|---|---|---|---|---|---|
| C. gracilis | ||||||
| CTL | 0.8% | 6.6% | 0.4% | 89.0% | 0.0% | 3.1% |
| Cu2+ | 1.1% | 7.4% | 0.5% | 87.9% | 0.0% | 3.0% |
| H2O2 | 0.8% | 11.2% | 0.4% | 83.4% | 0.0% | 4.2% |
| P. tricornutum | ||||||
| CTL | 65.5% | 4.0% | 1.7% | 28.2% | 0.0% | 0.6% |
| Cu2+ | 58.1% | 4.5% | 2.0% | 34.8% | 0.0% | 0.5% |
| H2O2 | 44.8% | 4.3% | 1.5% | 48.7% | 0.0% | 0.7% |
| T. lutea | ||||||
| CTL | 69.5% | 0.2% | 0.6% | 0.1% | 1.9% | 27.7% |
| Cu2+ | 73.6% | 0.3% | 0.8% | 0.1% | 2.2% | 23.0% |
| H2O2 | 67.9% | 0.2% | 0.7% | 0.1% | 1.7% | 29.4% |
| R. salina | ||||||
| CTL | 71.2% | 2.4% | 0.5% | 18.2% | 0.1% | 7.4% |
| Cu2+ | 66.7% | 2.7% | 0.4% | 21.4% | 0.2% | 8.6% |
| H2O2 | 79.1% | 3.5% | 1.5% | 12.4% | 0.1% | 3.4% |
| Component Name | CTL | Cu2+ | H2O2 | |||
|---|---|---|---|---|---|---|
| Conc. | sd | Conc. | sd | Conc. | sd | |
| 10-epi-10-F4t-NeuroP | 49.6 | 18.1 | 81.2 | 51.5 | 13.2 | 5.03 |
| 10-F4t-NeuroP | 40.0 | 11.9 | 62.5 | 34.3 | 15.1 | 5.99 |
| 13-epi-13-F4t-NeuroP | 113 | 35.7 | 160 | 76.8 | 39.2 | 13.4 |
| 13-F4t-NeuroP | 183 | 65.2 | 279 | 153 | 41.0 | NaN |
| 14(RS)-14-F4t-NeuroP | 51.0 | 14.8 | 89.0 | 55.0 | 12.3 | 4.78 |
| 15-epi-15-F2t-IsoP | 26.0 | 7.17 | 37.1 | 15.4 | 31.7 | 9.39 |
| 15-F2t-IsoP | 14.0 | 3.78 | 21.9 | 10.1 | 17.3 | 6.42 |
| 16-B1-PhytoP | 1960 | 96.9 | 2190 | 208 | 1410 | 844 |
| 18-F3t-IsoP | 711 | 239 | 1100 | 575 | 343 | 142 |
| 18-epi-18-F3t-IsoP | 240 | 63.6 | 393 | 203 | 174 | 54.9 |
| 20-epi-20-F4t-NeuroP | 67.0 | 19.7 | 97.2 | 48.0 | 37.3 | 16.2 |
| 20-F4t-NeuroP | 88.8 | 28.7 | 143.0 | 81.6 | 33.1 | 11.4 |
| 4(RS)-4-F3t-NeuroP | 13.4 | 4.57 | 22.7 | 16.8 | 9.24 | 1.90 |
| 4(RS)-4-F4t-NeuroP | 194 | 45.0 | 326 | 187 | 90.1 | 9.40 |
| 5-epi-5-F3t-IsoP | 457 | 137 | 717 | 378 | 278 | 77.1 |
| 5(RS)-5-F2t-IsoP | 70.1 | 17.7 | 107 | 49.0 | 95.7 | 22.3 |
| 5-F3t-IsoP | 424 | 107 | 713 | 377 | 193 | 43.3 |
| 5-F2c-IsoP | 149 | 31.0 | 222 | 96.2 | 143 | 34.9 |
| 7(RS)-ST-Δ18-11-dihomo-IsoF | 57.1 | 3.55 | 62.9 | 3.56 | 125 | 22.4 |
| 8-epi-8-F3t-IsoP | 38.9 | 11.5 | 61.4 | 33,0 | 18.9 | 7.81 |
| 8-F3t-IsoP | 57.3 | 17.0 | 82.0 | 40.2 | 17.0 | 4.21 |
| 9-epi-9-F1t-PhytoP | 514 | 115 | 851 | 455 | 668 | 119 |
| 9-F1t-PhytoP | 584 | 113 | 888 | 404 | 687 | 122 |
| 9-L1-PhytoP | 1510 | 76.6 | 1660 | 172 | 1110 | 619 |
| ent-16-epi-16-F1t-PhytoP | 440 | 96.7 | 715 | 362 | 550 | 118 |
| ent-16-F1t-PhytoP | 311 | 66.4 | 520 | 284 | 417 | 81.3 |
| ent-16(RS)-9-epi-ST-Δ14-10-PhytoF | 1790 | 173 | 2180 | 411 | 1290 | NaN |
| ent-9(RS)-12-epi-ST-Δ10-13-PhytoF | 434 | NaN | 570 | 124 | 404 | 149 |
| Component Name | CTL | Cu2+ | H2O2 | |||
|---|---|---|---|---|---|---|
| Conc. | sd | Conc. | sd | Conc. | sd | |
| 10-epi-10-F4t-NeuroP | 137 | 12.4 | 190 | NaN | 175 | 51.7 |
| 10-F4t-NeuroP | 105 | 10.3 | 153 | NaN | 132 | 37.5 |
| 13-epi-13-F4t-NeuroP | 282 | 15.4 | 312 | NaN | 334 | 71.2 |
| 13-F4t-NeuroP | 433 | 33.3 | 505 | NaN | 543 | 125 |
| 14(RS)-14-F4t-NeuroP | 132 | 15.8 | 159 | NaN | 167 | 72.9 |
| 16-B1-PhytoP | 988 | 99.0 | 1010 | NaN | 1050 | 169 |
| 16(RS)-16-A1-PhytoP | 324 | 30.2 | 587 | NaN | 348 | 39.3 |
| 18-F3t-IsoP | 4.54 | 1.13 | 4.60 | NaN | 2.46 | NaN |
| 20-epi-20-F4t-NeuroP | 116 | 15.3 | 213 | NaN | 144 | 33.2 |
| 20-F4t-NeuroP | 203 | 20.8 | 326 | NaN | 256 | 60.0 |
| 4(RS)-4-F3t-NeuroP | 135 | 17.5 | 261 | NaN | 138 | 15.3 |
| 4(RS)-4-F4t-NeuroP | 515 | 38.7 | 826 | NaN | 601 | 102 |
| 5(RS)-5-F2t-IsoP | 5.05 | 0.635 | 13.1 | NaN | 6.10 | 1.09 |
| 5-F2c-IsoP | 6.33 | NaN | 19.6 | NaN | 9.03 | 0.925 |
| 7(RS)-ST-Δ18-11-dihomo-IsoF | 41.8 | 3.50 | 78.5 | NaN | 49.5 | 12.2 |
| 8-epi-8-F3t-IsoP | 1.24 | 0.344 | 2.43 | NaN | 1.90 | 0.939 |
| 8-F3t-IsoP | 2.27 | 0.448 | 4.21 | NaN | 3.64 | 0.97 |
| 9-epi-9-F1t-PhytoP | 237 | 43.4 | 575 | NaN | 324 | 71.2 |
| 9-F1t-PhytoP | 407 | 33.6 | 730 | NaN | 466 | 65.9 |
| 9-L1-PhytoP | 727 | 82.2 | 1300 | NaN | 759 | 128 |
| ent-16-epi-16-F1t-PhytoP | 315 | 28.8 | 610 | NaN | 361 | 50.7 |
| ent-16-F1t-PhytoP | 381 | 20.4 | 682 | NaN | 409 | 66.6 |
| ent-16(RS)-13-epi-Δ14-9-PhytoF | 188 | 18.6 | 402 | NaN | 210 | 21.6 |
| ent-16(RS)-9-epi-ST-Δ14-10-PhytoF | 859 | 84.5 | 1810 | NaN | 1020 | 107 |
| ent-7(RS)-7-F2t-dihomo-IsoP | 3.26 | 0.602 | 13.7 | NaN | 4.47 | 1.48 |
| ent-9-D1t-PhytoP | 65.5 | 10.9 | 172 | NaN | 94.6 | 35.6 |
| ent-9-epi-9-D1t-PhytoP | 115 | 24.9 | 263 | NaN | 142 | 38.1 |
| ent-9(RS)-12-epi-ST-Δ10-13-PhytoF | 221 | 18.9 | 451 | NaN | 251 | 27.4 |
| Component Name | CTL | Cu2+ | H2O2 | |||
|---|---|---|---|---|---|---|
| Conc. | sd | Conc. | sd | Conc. | sd | |
| 10-epi-10-F4t-NeuroP | 5.24 | 0.437 | 6.23 | 1.32 | 5.68 | NaN |
| 10-F4t-NeuroP | 3.28 | 0.217 | 3.97 | 0.503 | 8.23 | 8.04 |
| 13-epi-13-F4t-NeuroP | 11.0 | 0.779 | 14.3 | 1.51 | 24.7 | 22.8 |
| 13-F4t-NeuroP | 14.6 | 1.88 | 16.6 | 3.60 | 31.8 | 25.8 |
| 15-epi-15-F2t-IsoP | 13.2 | 0.443 | 16 | 0.24 | 19.3 | 14.1 |
| 15-F2t-IsoP | 9.32 | 0.449 | 10.8 | 1.24 | 14.7 | 11.0 |
| 16-B1-PhytoP | 4.38 | 0.913 | 7.04 | 2.10 | 4.22 | 1.29 |
| 18-F3t-IsoP | 635 | 33.5 | 767 | NaN | 1040 | 792 |
| 18-epi-18-F3t-IsoP | 362 | 16.1 | 428 | NaN | 335 | NaN |
| 20-epi-20-F4t-NeuroP | 9.38 | 0.976 | 10.3 | NaN | 15.8 | 11.5 |
| 20-F4t-NeuroP | 10.7 | 2.15 | 10.3 | 2.79 | 17.7 | 13.4 |
| 4(RS)-4-F4t-NeuroP | 22.7 | 1.62 | 23.2 | 4.11 | 23.9 | NaN |
| 5-epi-5-F3t-IsoP | 603 | 26.6 | 661 | 92.3 | 569 | NaN |
| 5(RS)-5-F2t-IsoP | 35.1 | 3.19 | 41.4 | 7.19 | 61.1 | 52.8 |
| 5-F3t-IsoP | 471 | 22.2 | 477 | 80.4 | 493 | NaN |
| 5-F2c-IsoP | 105 | 3.71 | 140 | 20.1 | 248 | 180 |
| 7(RS)-ST-Δ18-11-dihomo-IsoF | 10.8 | 0.754 | 14.2 | 0.802 | 12.9 | 2.66 |
| 8-epi-8-F3t-IsoP | 57.5 | 3.01 | 63.7 | 7.63 | 56.0 | NaN |
| 8-F3t-IsoP | 52.4 | 2.05 | 62.9 | 7.24 | 56.6 | NaN |
| 9-F1t-PhytoP | 2.06 | 0.136 | 2.43 | 0.0889 | 2.83 | 1.29 |
| 9-L1-PhytoP | 3.21 | 0.587 | 5.40 | 1.53 | 3.27 | 0.92 |
| ent-16-epi-16-F1t-PhytoP | 1.44 | 0.164 | 1.84 | 0.229 | 2.04 | 0.952 |
| ent-16(RS)-9-epi-ST-Δ14-10-PhytoF | 4.44 | 0.354 | 7.08 | 1.33 | 5.51 | 1.59 |
| ent-9-epi-9-D1t-PhytoP | 3.96 | 0.347 | 6.51 | 1.02 | 7.54 | 5.75 |
| Component Name | CTL | Cu2+ | H2O2 | |||
|---|---|---|---|---|---|---|
| Conc. | sd | Conc. | sd | Conc. | sd | |
| 16-B1-PhytoP | 15.1 | 1.38 | 42.7 | 5.94 | 3.80 | 1.23 |
| 4(RS)-4-F4t-NeuroP | 1.84 | 0.0457 | 2.91 | 0.169 | 2.31 | 0.518 |
| 5-epi-5-F3t-IsoP | 47.1 | 2.74 | 99.9 | 14.0 | 86.6 | 49.4 |
| 5-F3t-IsoP | 33.7 | 2.07 | 72.9 | 8.36 | 57.9 | 32.4 |
| 5-F2c-IsoP | 13.0 | 1.02 | 25.2 | 3.05 | 14.2 | 2.78 |
| 7(RS)-ST-Δ18-11-dihomo-IsoF | 5.41 | 0.554 | 11.1 | 1.66 | 5.14 | 1.69 |
| 8-epi-8-F3t-IsoP | 6.72 | 0.759 | 12.0 | 1.83 | 10.8 | 4.19 |
| 8-F3t-IsoP | 3.92 | 0.304 | 7.99 | 0.911 | 6.92 | 3.48 |
| 9-F1t-PhytoP | 44.2 | 4.36 | 52.5 | 3.05 | 37.2 | 3.93 |
| 9-L1-PhytoP | 12.3 | 1.13 | 34.0 | 4.63 | 3.25 | 0.846 |
| ent-16-epi-16-F1t-PhytoP | 31.5 | 3.19 | 37.1 | 2.10 | 26.3 | 2.54 |
| ent-16-F1t-PhytoP | 81.0 | 8.05 | 90.9 | 4.69 | 67.3 | 7.15 |
| ent-16(RS)-13-epi-Δ14-9-PhytoF | 2.64 | 0.312 | 7.01 | NaN | 1.06 | 0.268 |
| ent-16(RS)-9-epi-ST-Δ14-10-PhytoF | 20.2 | 2.46 | 44.0 | 4.61 | 5.78 | 1.94 |
| ent-9-epi-9-D1t-PhytoP | 0.918 | 0.182 | 2.40 | 0.392 | 0.783 | 0.426 |
| ent-9(RS)-12-epi-ST-Δ10-13-PhytoF | 4.53 | 0.744 | 11.7 | 1.65 | 3.58 | 0.882 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vigor, C.; Oger, C.; Reversat, G.; Rocher, A.; Zhou, B.; Linares-Maurizi, A.; Guy, A.; Bultel-Poncé, V.; Galano, J.-M.; Vercauteren, J.; et al. Isoprostanoid Profiling of Marine Microalgae. Biomolecules 2020, 10, 1073. https://doi.org/10.3390/biom10071073
Vigor C, Oger C, Reversat G, Rocher A, Zhou B, Linares-Maurizi A, Guy A, Bultel-Poncé V, Galano J-M, Vercauteren J, et al. Isoprostanoid Profiling of Marine Microalgae. Biomolecules. 2020; 10(7):1073. https://doi.org/10.3390/biom10071073
Chicago/Turabian StyleVigor, Claire, Camille Oger, Guillaume Reversat, Amandine Rocher, Bingqing Zhou, Amandyne Linares-Maurizi, Alexandre Guy, Valérie Bultel-Poncé, Jean-Marie Galano, Joseph Vercauteren, and et al. 2020. "Isoprostanoid Profiling of Marine Microalgae" Biomolecules 10, no. 7: 1073. https://doi.org/10.3390/biom10071073
APA StyleVigor, C., Oger, C., Reversat, G., Rocher, A., Zhou, B., Linares-Maurizi, A., Guy, A., Bultel-Poncé, V., Galano, J. -M., Vercauteren, J., Durand, T., Potin, P., Tonon, T., & Leblanc, C. (2020). Isoprostanoid Profiling of Marine Microalgae. Biomolecules, 10(7), 1073. https://doi.org/10.3390/biom10071073